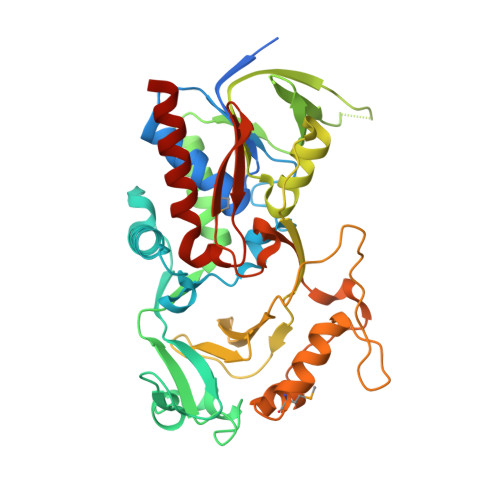
Crystal structure of DR_0571 protein from Deinococcus radiodurans in complex with ADP.
Forouhar, F., Chen, Y., Seetharaman, J., Mao, L., Xiao, R., Cunningham, K., Owen, L.A., Maglaqui, M., Baran, M.C., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.