Inhibition of lactoperoxidase by its own catalytic product: crystal structure of the hypothiocyanate-inhibited bovine lactoperoxidase at 2.3-A resolution.
Singh, A.K., Singh, N., Sharma, S., Shin, K., Takase, M., Kaur, P., Srinivasan, A., Singh, T.P.(2009) Biophys J 96: 646-654
- PubMed: 19167310
- DOI: https://doi.org/10.1016/j.bpj.2008.09.019
- Primary Citation of Related Structures:
3BXI - PubMed Abstract:
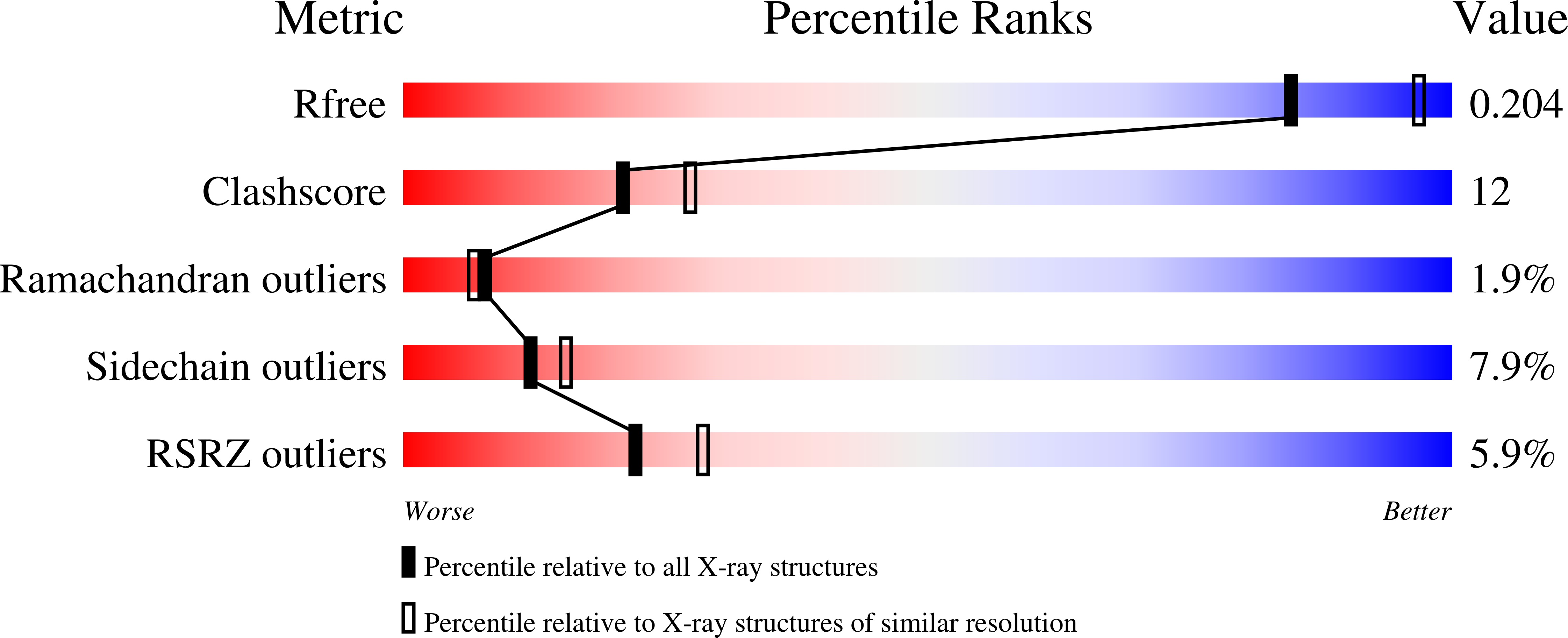
To the best of our knowledge, this is the first report on the structure of product-inhibited mammalian peroxidase. Lactoperoxidase is a heme containing an enzyme that catalyzes the inactivation of a wide range of microorganisms. In the presence of hydrogen peroxide, it preferentially converts thiocyanate ion into a toxic hypothiocyanate ion. Samples of bovine lactoperoxidase containing thiocyanate (SCN(-)) and hypothiocyanate (OSCN(-)) ions were purified and crystallized. The structure was determined at 2.3-A resolution and refined to R(cryst) and R(free) factors of 0.184 and 0.221, respectively. The determination of structure revealed the presence of an OSCN(-) ion at the distal heme cavity. The presence of OSCN(-) ions in crystal samples was also confirmed by chemical and spectroscopic analysis. The OSCN(-) ion interacts with the heme iron, Gln-105 N(epsilon1), His-109 N(epsilon2), and a water molecule W96. The sulfur atom of the OSCN(-) ion forms a hypervalent bond with a nitrogen atom of the pyrrole ring D of the heme moiety at an S-N distance of 2.8 A. The heme group is covalently bound to the protein through two ester linkages involving carboxylic groups of Glu-258 and Asp-108 and the modified methyl groups of pyrrole rings A and C, respectively. The heme moiety is significantly distorted from planarity, whereas pyrrole rings A, B, C, and D are essentially planar. The iron atom is displaced by approximately 0.2 A from the plane of the heme group toward the proximal site. The substrate channel resembles a long tunnel whose inner walls contain predominantly aromatic residues such as Phe-113, Phe-239, Phe-254, Phe-380, Phe-381, Phe-422, and Pro-424. A phosphorylated Ser-198 was evident at the surface, in the proximity of the calcium-binding channel.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.