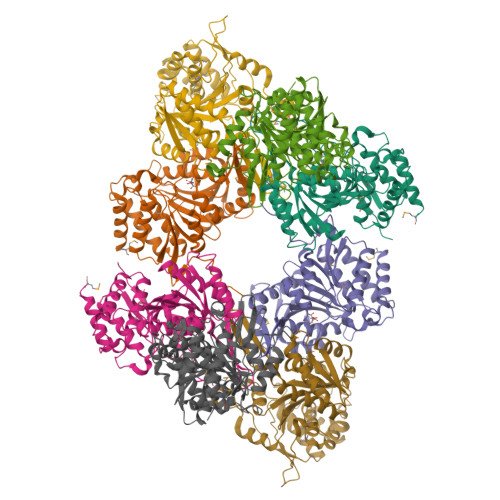
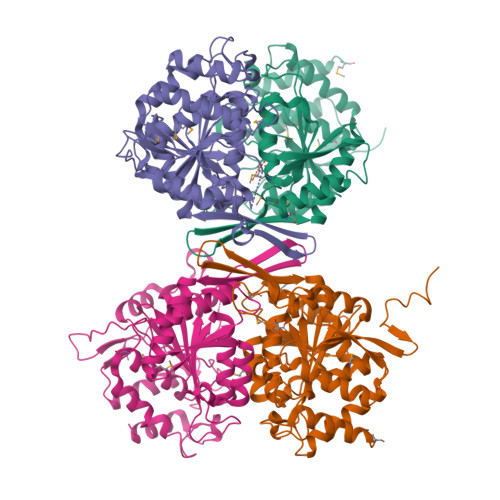
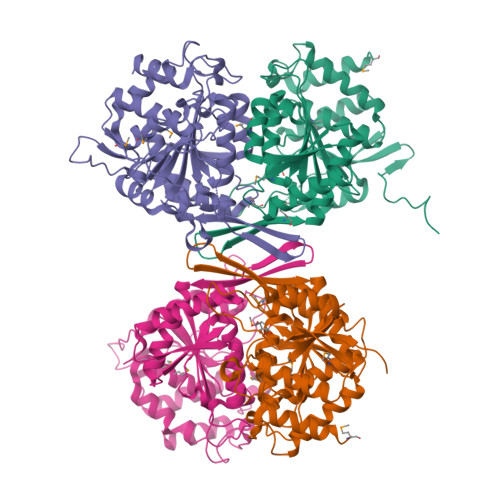
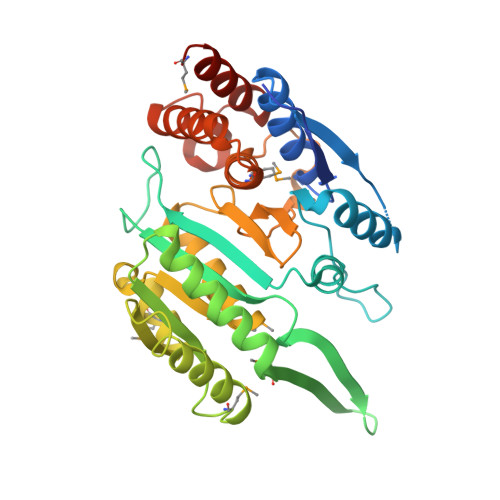
Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
Taylor, A.B., Hu, G., Hart, P.J., McAlister-Henn, L.(2008) J Biological Chem 283: 10872-10880
- PubMed: 18256028
- DOI: https://doi.org/10.1074/jbc.M708719200
- Primary Citation of Related Structures:
3BLV, 3BLW, 3BLX - PubMed Abstract:
Mitochondrial NAD(+)-specific isocitrate dehydrogenases (IDHs) are key regulators of flux through biosynthetic and oxidative pathways in response to cellular energy levels. Here we present the first structures of a eukaryotic member of this enzyme family, the allosteric, hetero-octameric, NAD(+)-specific IDH from yeast in three forms: 1) without ligands, 2) with bound analog citrate, and 3) with bound citrate + AMP. The structures reveal the molecular basis for ligand binding to homologous but distinct regulatory and catalytic sites positioned at the interfaces between IDH1 and IDH2 subunits and define pathways of communication between heterodimers and heterotetramers in the hetero-octamer. Disulfide bonds observed at the heterotetrameric interfaces in the unliganded IDH hetero-octamer are reduced in the ligand-bound forms, suggesting a redox regulatory mechanism that may be analogous to the "on-off" regulation of non-allosteric bacterial IDHs via phosphorylation. The results strongly suggest that eukaryotic IDH enzymes are exquisitely tuned to ensure that allosteric activation occurs only when concentrations of isocitrate are elevated.
Organizational Affiliation:
Department of Biochemistry and X-ray Crystallography Core Laboratory, The University of Texas Health Science Center, San Antonio, Texas 78229, USA.