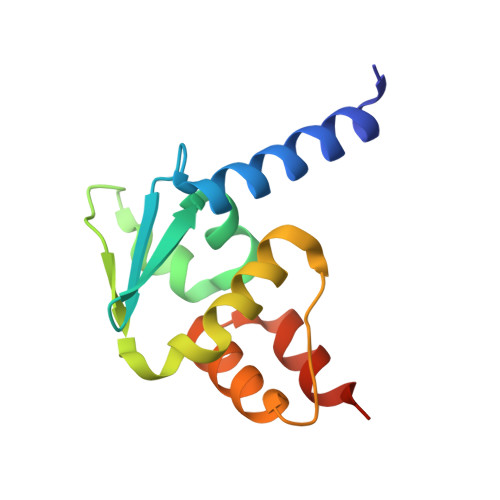
Crystal Structure of the Human BTB domain of the Krueppel related Zinc Finger Protein 3 (HKR3).
Filippakopoulos, P., Bullock, A., Cooper, C., Keates, T., Salah, E., Pilka, E., Pike, A.C.W., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Weigelt, J., Knapp, S.To be published.