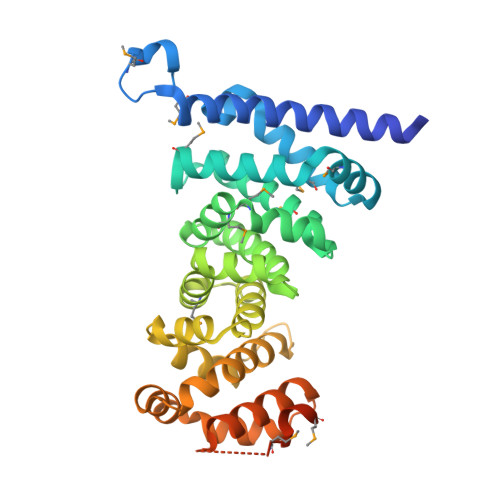
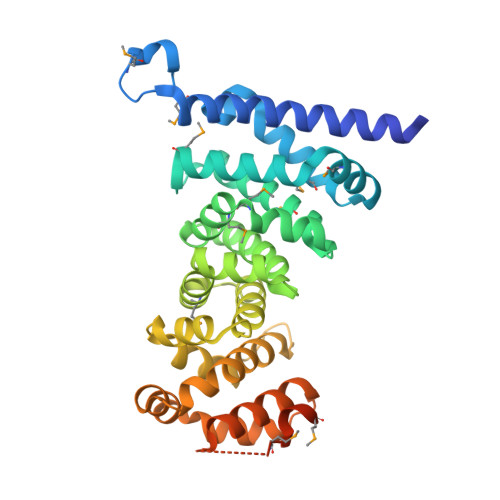
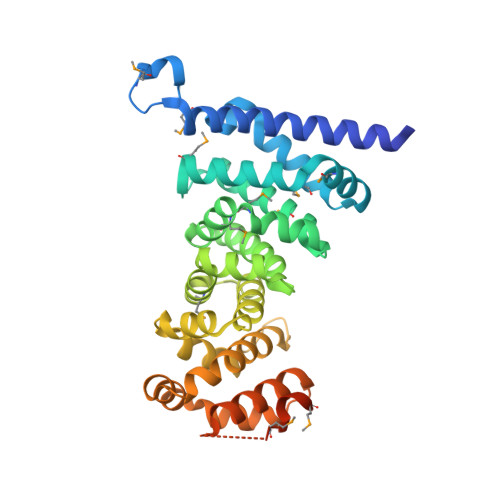
Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Morishita, E.C., Murayama, K., Kato-Murayama, M., Ishizuka-Katsura, Y., Tomabechi, Y., Hayashi, T., Terada, T., Handa, N., Shirouzu, M., Akiyama, T., Yokoyama, S.(2011) Structure 19: 1496-1508
- PubMed: 22000517
- DOI: https://doi.org/10.1016/j.str.2011.07.013
- Primary Citation of Related Structures:
3AU3, 3QHE - PubMed Abstract:
Adenomatous polyposis coli (APC) is a tumor suppressor protein commonly mutated in colorectal tumors. APC plays important roles in Wnt signaling and other cellular processes. Here, we present the crystal structure of the armadillo repeat (Arm) domain of APC, which facilitates the binding of APC to various proteins. APC-Arm forms a superhelix with a positively charged groove. We also determined the structure of the complex of APC-Arm with the tyrosine-rich (YY) domain of the Src-associated in mitosis, 68 kDa protein (Sam68), which regulates TCF-1 alternative splicing. Sam68-YY forms numerous interactions with the residues on the groove and is thereby fixed in a bent conformation. We assessed the effects of mutations and phosphorylation on complex formation between APC-Arm and Sam68-YY. Structural comparisons revealed different modes of ligand recognition between the Arm domains of APC and other Arm-containing proteins.
Organizational Affiliation:
RIKEN Systems and Structural Biology Center, 1-7-22 Suehiro-cho, Tsurumi, Yokohama 230-0045, Japan.