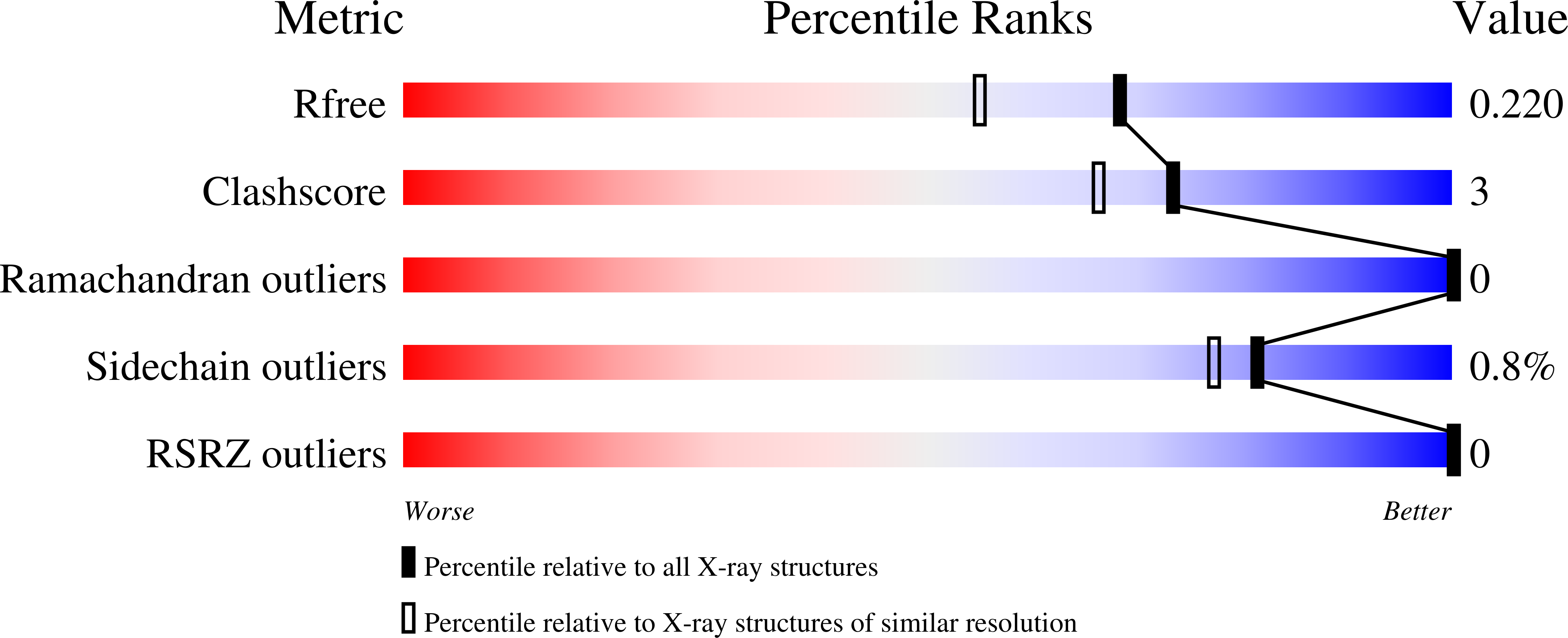
Involvement of the distal Arg residue in Cl- binding of midge larval haemoglobin
Kuwada, T., Hasegawa, T., Takagi, T., Sakae, T., Sato, I., Shishikura, F.(2011) Acta Crystallogr D Biol Crystallogr 67: 488-495
- PubMed: 21543852
- DOI: https://doi.org/10.1107/S0907444911010808
- Primary Citation of Related Structures:
3ARJ, 3ARK, 3ARL - PubMed Abstract:
Monomeric haemoglobin component V (Hb V) from the larva of the midge Propsilocerus akamusi shows high Cl⁻ affinity under high salt concentrations at acidic pH. In order to understand the structural changes that depend on Cl⁻ binding, crystal structures of Hb V were determined under acidic high-salt conditions and the structural changes arising from different haem-bound ligands were simulated. Crystal structures of Hb V under acidic high-salt conditions indicated that the side chain of ArgE10 on the distal face of the haem contributes to stabilizing haem-bound Cl⁻. The conformation of the Arg side chain in the Cl⁻-bound form was almost identical to that in ligated Hb V at neutral pH but not to that in met Hb V under acidic salt-free conditions. Furthermore, preliminary molecular-dynamics simulations also indicated that the swinging of the Arg side chain into the haem pocket depends on Cl⁻ ligation. This result suggests that, like pH change, Cl⁻ binding affects the location of the distal Arg residue. Owing to the increased positive electrostatic potential observed in the haem pocket at acidic pH, it was concluded that electrostatic changes caused by pH change and anionic ligand binding may affect the behaviour of the polar Arg residue.
Organizational Affiliation:
Laboratory for Electron Beam Research and Application (LEBRA), Institute of Quantum Science, Nihon University, Funabashi, Chiba 274-8501, Japan. kuwadat@lebra.nihon-u.ac.jp