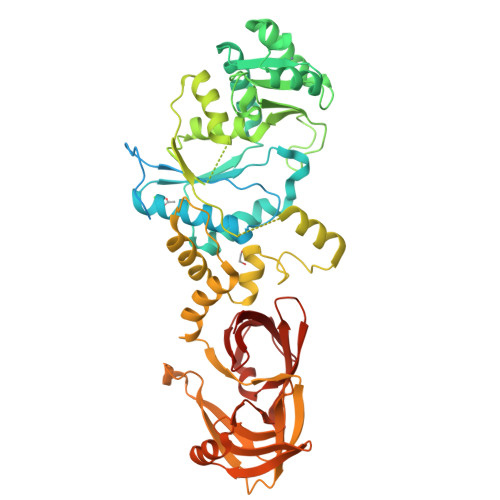
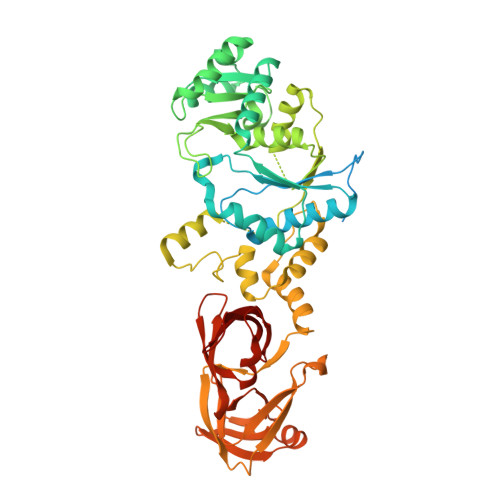
Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: a missing link in the evolution of Gln-tRNAGln formation
Nureki, O., O'Donoghue, P., Watanabe, N., Ohmori, A., Oshikane, H., Araiso, Y., Sheppard, K., Soll, D., Ishitani, R.(2010) Nucleic Acids Res
- PubMed: 20601684
- DOI: https://doi.org/10.1093/nar/gkq605
- Primary Citation of Related Structures:
3AII - PubMed Abstract:
The molecular basis of the genetic code relies on the specific ligation of amino acids to their cognate tRNA molecules. However, two pathways exist for the formation of Gln-tRNA(Gln). The evolutionarily older indirect route utilizes a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) that can form both Glu-tRNA(Glu) and Glu-tRNA(Gln). The Glu-tRNA(Gln) is then converted to Gln-tRNA(Gln) by an amidotransferase. Since the well-characterized bacterial ND-GluRS enzymes recognize tRNA(Glu) and tRNA(Gln) with an unrelated α-helical cage domain in contrast to the β-barrel anticodon-binding domain in archaeal and eukaryotic GluRSs, the mode of tRNA(Glu)/tRNA(Gln) discrimination in archaea and eukaryotes was unknown. Here, we present the crystal structure of the Methanothermobacter thermautotrophicus ND-GluRS, which is the evolutionary predecessor of both the glutaminyl-tRNA synthetase (GlnRS) and the eukaryotic discriminating GluRS. Comparison with the previously solved structure of the Escherichia coli GlnRS-tRNA(Gln) complex reveals the structural determinants responsible for specific tRNA(Gln) recognition by GlnRS compared to promiscuous recognition of both tRNAs by the ND-GluRS. The structure also shows the amino acid recognition pocket of GluRS is more variable than that found in GlnRS. Phylogenetic analysis is used to reconstruct the key events in the evolution from indirect to direct genetic encoding of glutamine.
Organizational Affiliation:
Department of Basic Medical Sciences, Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan. nureki@ims.u-tokyo.ac.jp