X-ray crystal structure of the light-independent protochlorophyllide reductase
Muraki, N., Nomata, J., Ebata, K., Mizoguchi, T., Shiba, T., Tamiaki, H., Kurisu, G., Fujita, Y.(2010) Nature 465: 110-114
- PubMed: 20400946
- DOI: https://doi.org/10.1038/nature08950
- Primary Citation of Related Structures:
3AEK, 3AEQ, 3AER, 3AES, 3AET, 3AEU - PubMed Abstract:
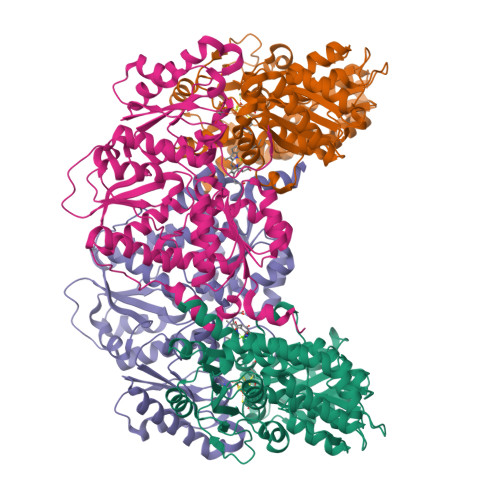
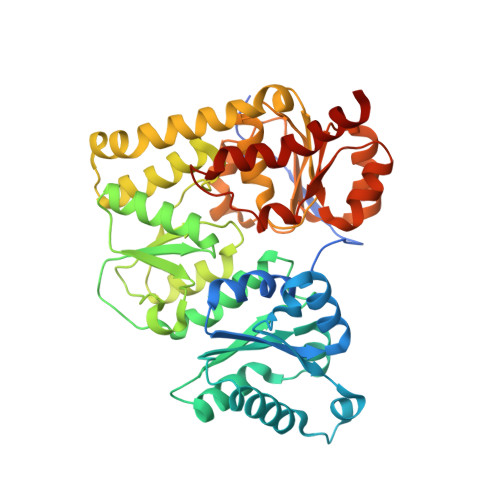
Photosynthetic organisms adopt two different strategies for the reduction of the C17 = C18 double bond of protochlorophyllide (Pchlide) to form chlorophyllide a, the direct precursor of chlorophyll a (refs 1-4). The first involves the activity of the light-dependent Pchlide oxidoreductase, and the second involves the light-independent (dark-operative) Pchlide oxidoreductase (DPOR). DPOR is a nitrogenase-like enzyme consisting of two components, L-protein (a BchL dimer) and NB-protein (a BchN-BchB heterotetramer), which are structurally related to nitrogenase Fe protein and MoFe protein, respectively. Here we report the crystal structure of the NB-protein of DPOR from Rhodobacter capsulatus at a resolution of 2.3A. As expected, the overall structure is similar to that of nitrogenase MoFe protein: each catalytic BchN-BchB unit contains one Pchlide and one iron-sulphur cluster (NB-cluster) coordinated uniquely by one aspartate and three cysteines. Unique aspartate ligation is not necessarily needed for the cluster assembly but is essential for the catalytic activity. Specific Pchlide-binding accompanies the partial unwinding of an alpha-helix that belongs to the next catalytic BchN-BchB unit. We propose a unique trans-specific reduction mechanism in which the distorted C17-propionate of Pchlide and an aspartate from BchB serve as proton donors for C18 and C17 of Pchlide, respectively. Intriguingly, the spatial arrangement of the NB-cluster and Pchlide is almost identical to that of the P-cluster and FeMo-cofactor in nitrogenase MoFe-protein, illustrating that a common architecture exists to reduce chemically stable multibonds of porphyrin and dinitrogen.
Organizational Affiliation:
Department of Life Sciences, University of Tokyo, Komaba, Meguro-ku, Tokyo 153-8902, Japan.