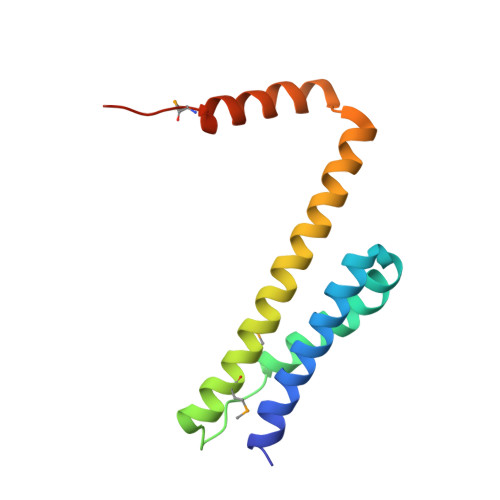
Structural insight into the regulatory mechanisms of interactions of the flagellar type III chaperone FliT with its binding partners.
Imada, K., Minamino, T., Kinoshita, M., Furukawa, Y., Namba, K.(2010) Proc Natl Acad Sci U S A 107: 8812-8817
- PubMed: 20421493
- DOI: https://doi.org/10.1073/pnas.1001866107
- Primary Citation of Related Structures:
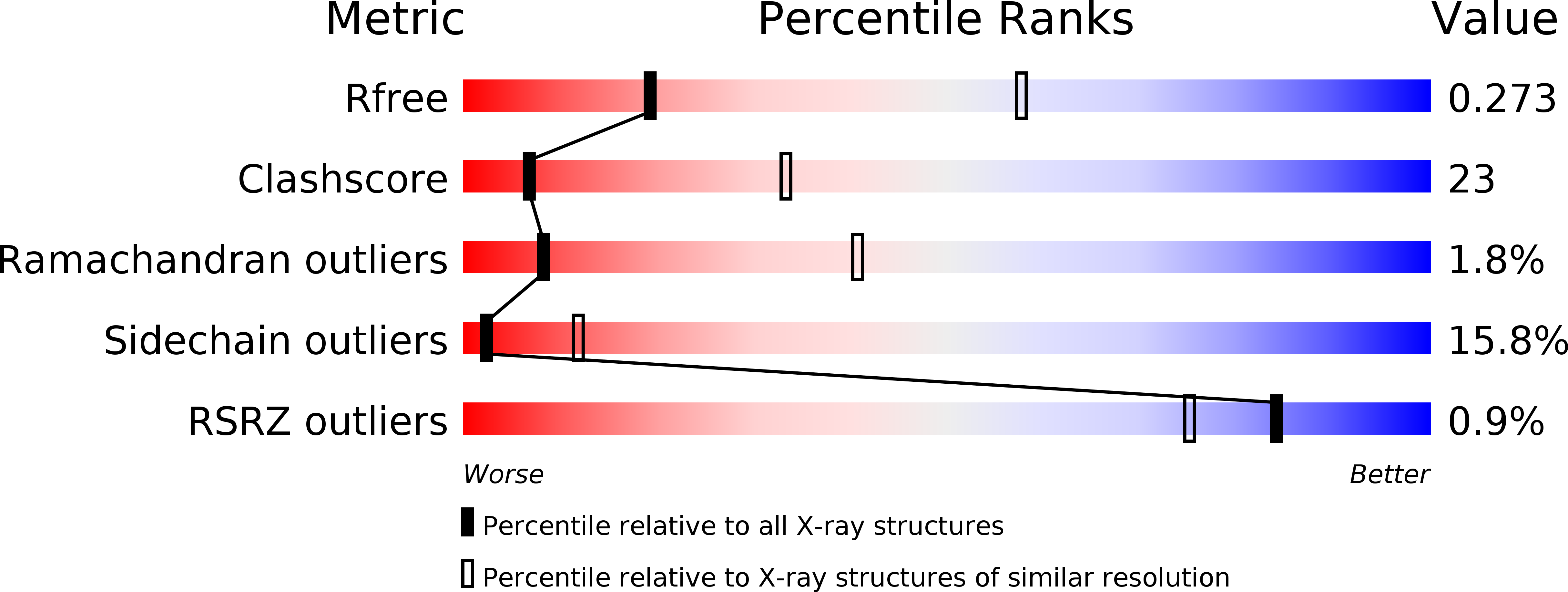
3A7M - PubMed Abstract:
For self-assembly of the bacterial flagellum, most of the flagellar component proteins synthesized in the cytoplasm are exported by the flagellar type III export apparatus to the growing, distal end. Flagellar protein export is highly organized and well controlled in every step of the flagellar assembly process. Flagellar-specific chaperones not only facilitate the export of their cognate proteins, as well as prevent their premature aggregation in the cytoplasm, but also play a role in fine-tuning flagellar gene expression to be coupled with the flagellar assembly process. FliT is a flagellar-specific chaperone responsible for the export of the filament-capping protein FliD and for negative control of flagellar gene expression by binding to the FlhDC complex. Here we report the crystal structure of Salmonella FliT at 3.2-A resolution. The structural and biochemical analyses clearly reveal that the C-terminal segment of FliT regulates its interactions with the FlhDC complex, FliI ATPase, and FliJ (subunits of the export apparatus), and that its conformational change is responsible for the switch in its binding partners during flagellar protein export.
Organizational Affiliation:
Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan. kimada@fbs.osaka-u.ac.jp