Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis
Vivian, J.P., Scoullar, J., Rimmer, K., Bushell, S.R., Beddoe, T., Wilce, M.C.J., Byres, E., Boyle, T.P., Doak, B., Simpson, J.S., Graham, B., Heras, B., Kahler, C.M., Rossjohn, J., Scanlon, M.J.(2009) J Mol Biology 394: 931-943
- PubMed: 19815019
- DOI: https://doi.org/10.1016/j.jmb.2009.09.065
- Primary Citation of Related Structures:
3A3T - PubMed Abstract:
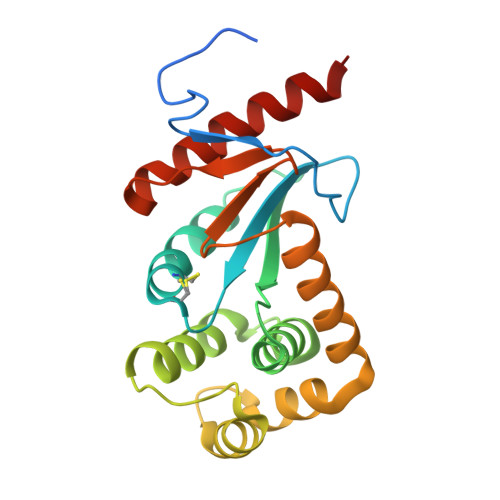
Neisseria meningitidis encodes three DsbA oxidoreductases (NmDsbA1-NmDsbA3) that are vital for the oxidative folding of many membrane and secreted proteins, and these three enzymes are considered to exhibit different substrate specificities. This has led to the suggestion that each N. meningitidis DsbA (NmDsbA) may play a specialized role in different stages of pathogenesis; however, the molecular and structural bases of the different roles of NmDsbAs are unclear. With the aim of determining the molecular basis for substrate specificity and how this correlates to pathogenesis, we undertook a biochemical and structural characterization of the three NmDsbAs. We report the 2.0-A-resolution crystal structure of the oxidized form of NmDsbA1, which adopted a canonical DsbA fold similar to that observed in the structures of NmDsbA3 and Escherichia coli DsbA (EcDsbA). Structural comparisons revealed variations around the active site and candidate peptide-binding region. Additionally, we demonstrate that all three NmDsbAs are strong oxidases with similar redox potentials; however, they differ from EcDsbA in their ability to be reoxidized by E. coli DsbB. Collectively, our studies suggest that the small structural differences between the NmDsbA enzymes and EcDsbA are functionally significant and are the likely determinants of substrate specificity.
Organizational Affiliation:
The Protein Crystallography Unit, Australian Research Council Center of Excellence in Structural and Functional Microbial Genomics, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.