Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
Ding, J., Bao, J., Zhu, D., Zhang, Y., Wang, D.C.(2010) J Struct Biol 171: 309-317
- PubMed: 20546901
- DOI: https://doi.org/10.1016/j.jsb.2010.05.009
- Primary Citation of Related Structures:
3A0C, 3A0D, 3A0E - PubMed Abstract:
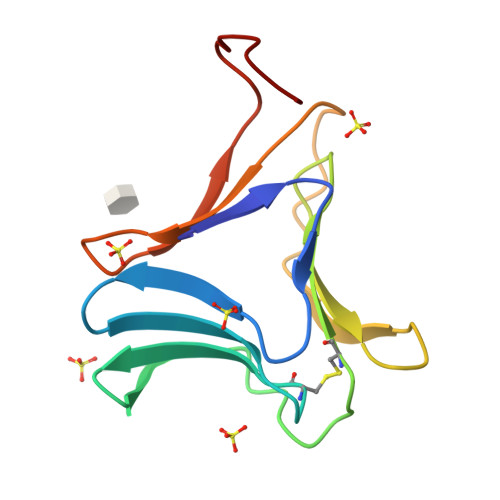
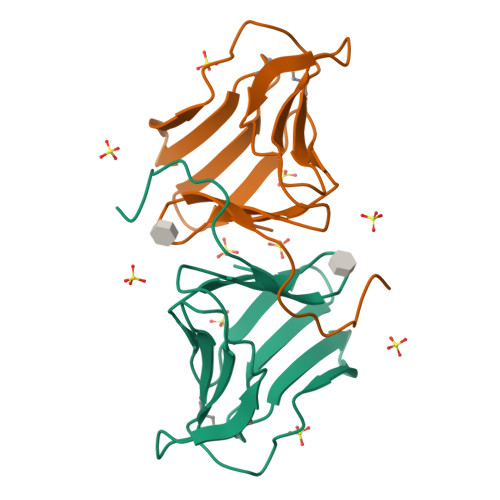
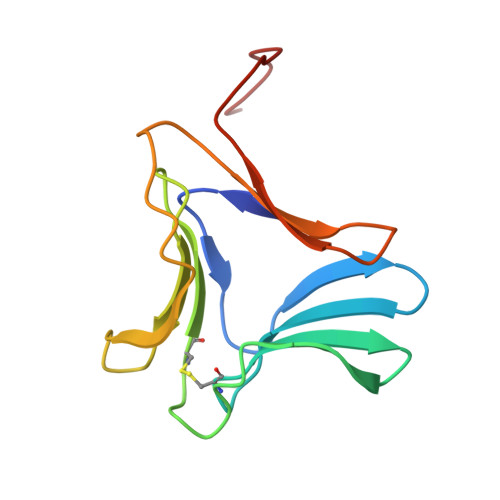
Polygonatum cyrtonema lectin (PCL) is a novel anti-HIV mannose-binding lectin from Galanthus nivalis agglutinin (GNA)-related lectin family. Crystal structures of ligand-free PCL and its complexes with monomannoside and alpha1-3 dimannoside have been determined. The ligand-free PCL is dimeric, with both subunits adopt the beta-prism II fold. PCL subunit binds mannose using a potential bivalent mode instead of the usual trivalent mode, in which carbohydrate-binding site (CBS) I and CBS III adopt the conserved mannose-binding motif of QXDXNXVXY (X is one of any amino acid residues) as observed in other structurally characterized GNA-related lectins, while CBS II adopts a modified motif with residues Gln58 and Asp60, which are critical for mannose-binding, substituted by His58 and Asn60, respectively. As a result, CBS II is unfit for mannose-binding. In the mannoside complexes, ligand-bindings only occur at CBS I which provides the specificity for alpha1-3 dimannoside. CBS II and CBS III are cooperatively occupied by a well-ordered sulfate ion, through which the individual dimers are cross-linked to form a unique super-structure of 3(2) helical lattice. Surveying the sequences of GNA-related lectins revealed that the modified binding motif of CBS II is widely distributed in the Liliaceae family as an intrinsic structural element. There is evidence that other GNA-related lectins will also adopt the similar super-structure as PCL. Thus PCL structure, unique in ligand-binding mode, may represent a novel type of structure of GNA-related lectins. Comparative analyses indicated that the dimer-based super-structure may play a primary role in the anti-HIV property of PCL.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People's Republic of China.