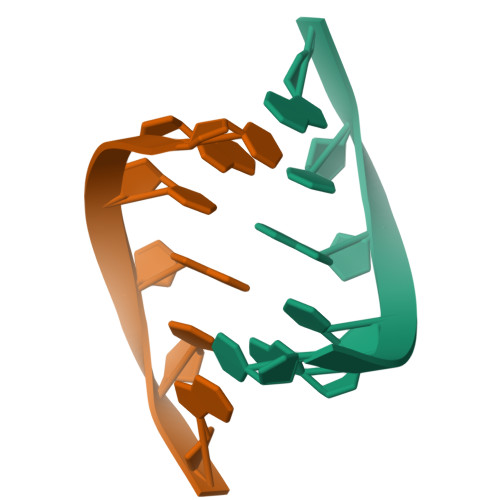
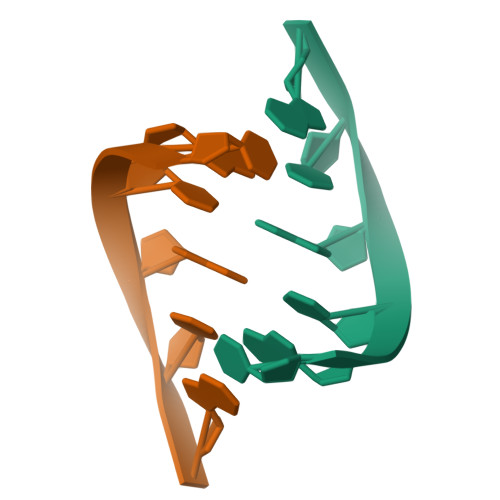
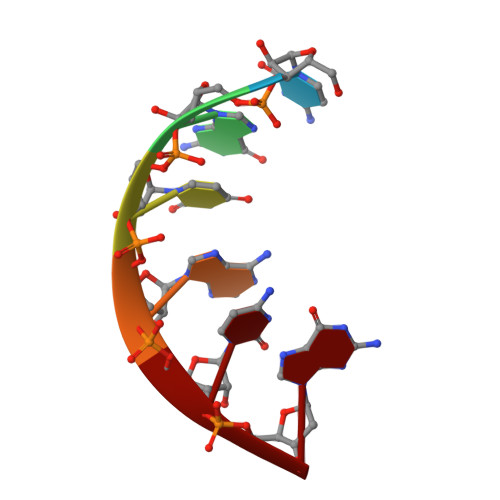
1.76 A structure of a pyrimidine start alternating A-RNA hexamer r(CGUAC)dG.
Biswas, R., Mitra, S.N., Sundaralingam, M.(1998) Acta Crystallogr D Biol Crystallogr 54: 570-576
- PubMed: 9761851
- DOI: https://doi.org/10.1107/s0907444997013097
- Primary Citation of Related Structures:
377D - PubMed Abstract:
The crystal structure of the alternating RNA r(CGUAC)dG with a 3'-terminal deoxy G residue has been determined at 1.76 A resolution. The crystal belongs to the orthorhombic space group C2221, unit-cell dimensions a = 29.53, b = 44.61 and c = 94.18 A, with two independent duplexes (I and II) per asymmetric unit. The structure was solved by the molecular-replacement method. The final R factor was 18.8% using 4757 reflections in the resolution range 8.0-1.76 A. The model contains a total of 496 atoms and 85 solvent molecules. The two duplexes form the repeating unit and stack in the usual head-to-tail (5',3'/5',3') fashion into a pseudocontinuous helical column. Almost all of the 2'-hydroxyl groups are engaged in the three modes of water-mediated interactions to the base N3/O2 atoms, the sugar O4' atoms and the backbone phosphates. Thus, the 2'-hydroxyl group of RNA is probably contributing to the stability of the duplexes.
Organizational Affiliation:
Biological Macromolecular Structure Center, Departments of Chemistry, Biochemistry and The Ohio State Biochemistry Program, The Ohio State University, 100 West 18th Avenue, Columbus, OH 43210, USA.