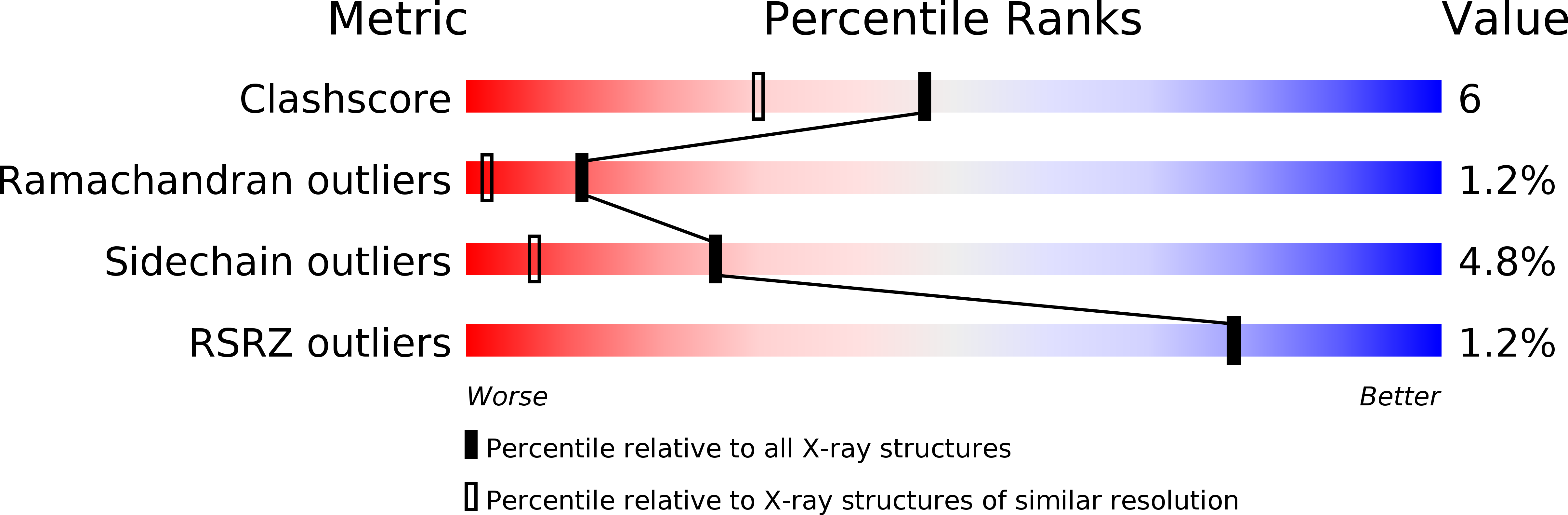
Structure of cytochrome c551 from Pseudomonas aeruginosa refined at 1.6 A resolution and comparison of the two redox forms.
Matsuura, Y., Takano, T., Dickerson, R.E.(1982) J Mol Biol 156: 389-409
- PubMed: 6283101
- DOI: https://doi.org/10.1016/0022-2836(82)90335-7
- Primary Citation of Related Structures:
351C, 451C