Crystallographic evidence of drastic movement of a copper ion toward the substrate tyrosine for starting hydroxylation reaction of tyrosinase
Matoba, Y., Yoshitsu, H., Jeon, H.J., Oda, K., Noda, M., Kumagai, T., Sugiyama, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tyrosinase | 281 | Streptomyces castaneoglobisporus | Mutation(s): 0 EC: 1.14.18.1 | 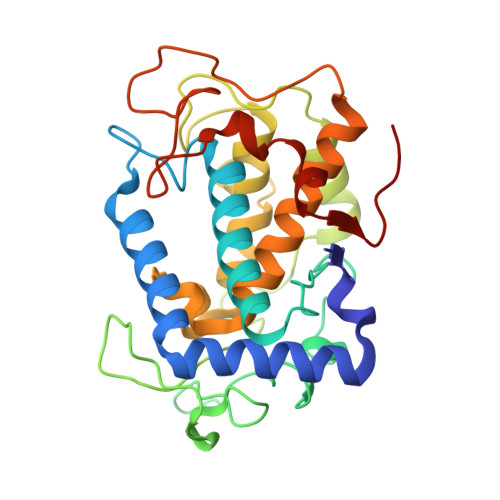 | |
UniProt | |||||
Find proteins for Q83WS2 (Streptomyces castaneoglobisporus) Explore Q83WS2 Go to UniProtKB: Q83WS2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q83WS2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MelC | 134 | Streptomyces castaneoglobisporus | Mutation(s): 0 | 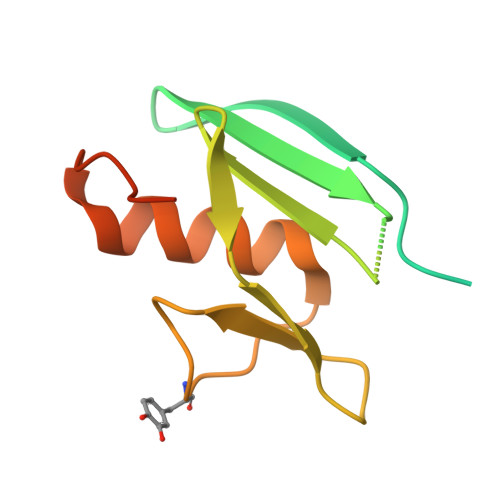 | |
UniProt | |||||
Find proteins for Q83WS1 (Streptomyces castaneoglobisporus) Explore Q83WS1 Go to UniProtKB: Q83WS1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q83WS1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CU Query on CU | C [auth A], D [auth A], E [auth A], J [auth B] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| NO3 Query on NO3 | F [auth A], G [auth A], H [auth A], I [auth A], K [auth B] | NITRATE ION N O3 NHNBFGGVMKEFGY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| DAH Query on DAH | B | L-PEPTIDE LINKING | C9 H11 N O4 |  | TYR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 65.16 | α = 90 |
| b = 97.68 | β = 90 |
| c = 54.99 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| SHELXL-97 | refinement |
| BSS | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |