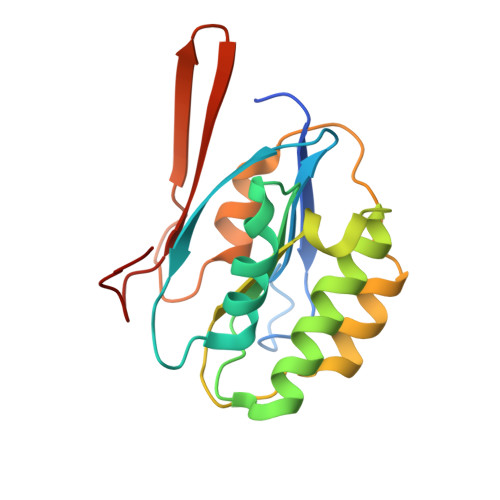
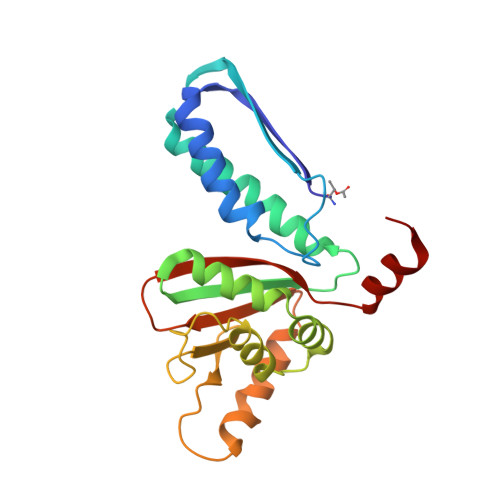
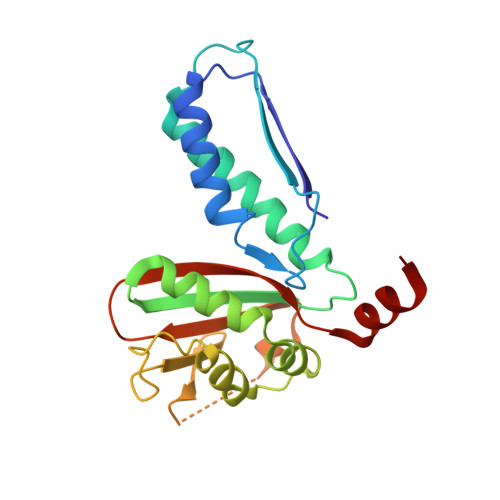
Structural and Biochemical Analyses Reveal How Ornithine Acetyl Transferase Binds Acidic and Basic Amino Acid Substrates.
Iqbal, A., Clifton, I.J., Chowdhury, R., Ivison, D., Domene, C., Schofield, C.J.(2011) Org Biomol Chem 9: 6219
- PubMed: 21796301
- DOI: https://doi.org/10.1039/c1ob05554b
- Primary Citation of Related Structures:
2YEP - PubMed Abstract:
Structural and biochemical analyses reveal how ornithine acetyl-transferases catalyse the reversible transfer of an acetyl-group from a basic (ornithine) to an acidic (glutamate) amino acid by employing a common mechanism involving an acetyl-enzyme intermediate but using different side chain binding modes.
- Chemistry Research Laboratory, Department of Chemistry, University of Oxford, Oxford, UK.
Organizational Affiliation: