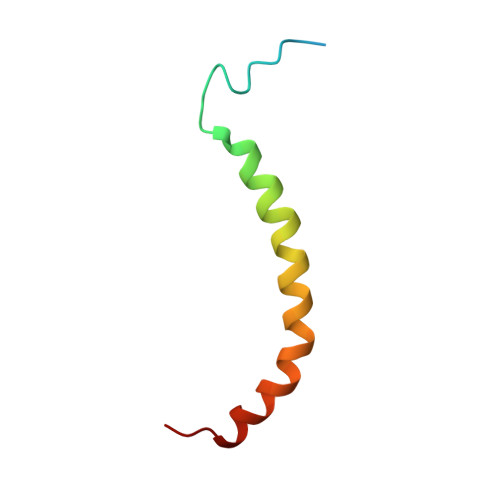
Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biukovic, G., Gayen, S., Pervushin, K., Gruber, G.(2009) Biophys J 97: 286-294
- PubMed: 19580766
- DOI: https://doi.org/10.1016/j.bpj.2009.04.026
- Primary Citation of Related Structures:
2K6I - PubMed Abstract:
A series of truncated forms of subunit H were generated to establish the domain features of that protein. Circular dichroism analysis demonstrated that H is divided at least into a C-terminal coiled-coil domain within residues 54-104, and an N-terminal domain formed by adjacent alpha-helices. With a cysteine at the C-terminus of each of the truncated proteins (H(1-47), H(1-54), H(1-59), H(1-61), H(1-67), H(1-69), H(1-71), H(1-78), H(1-80), H(1-91), and H(47-105)), the residues involved in formation of the coiled-coil interface were determined. Proteins H(1-54), H(1-61), H(1-69), and H(1-80) showed strong cross-link formation, which was weaker in H(1-47), H(1-59), H(1-71), and H(1-91). A shift in disulfide formation between cysteines at positions 71 and 80 reflected an interruption in the periodicity of hydrophobic residues in the region 71AEKILEETEKE81. To understand how the N-terminal domain of H is formed, we determined for the first time, to our knowledge, the solution NMR structure of H(1-47), which revealed an alpha-helix between residues 15-42 and a flexible N-terminal stretch. The alpha-helix includes a kink that would bring the two helices of the C-terminus into the coiled-coil arrangement. H(1-47) revealed a strip of alanines involved in dimerization, which were tested by exchange to single cysteines in subunit H mutants.
- School of Biological Sciences, Nanyang Technological University, Singapore.
Organizational Affiliation: