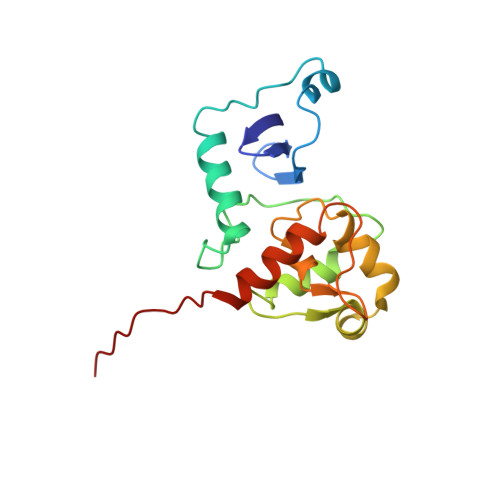
Structural studies of MJ1529, an O(6)-methylguanine-DNA methyltransferase
Roberts, A., Pelton, J.G., Wemmer, D.E.(2006) Magn Reson Chem 44: 71-82
- PubMed: 16826543
- DOI: https://doi.org/10.1002/mrc.1823
- Primary Citation of Related Structures:
2G7H - PubMed Abstract:
The structure of an O6-methylguanine-DNA methyltransferase (MGMT) from the thermophile Methanococcus jannaschii has been determined using multinuclear multidimensional NMR spectroscopy. The structure is similar to homologs from other organisms that have been determined by crystallography, with some variation in the N-terminal domain. The C-terminal domain is more highly conserved in both sequence and structure. Regions of the protein show broadening, reflecting conformational flexibility that is likely related to function.
- Department of Chemistry, University of California and Physical Biosciences Division, Lawrence Berkeley National Lab, Berkeley, CA 94720-1460, USA.
Organizational Affiliation: