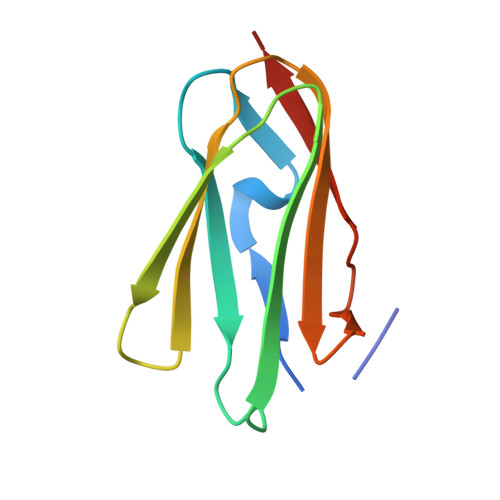
The Structure of the Gpib-Filamin a Complex.
Nakamura, F., Pudas, R., Heikkinen, O., Permi, P., Kilpelainen, I., Munday, A.D., Hartwig, J.H., Stossel, T.P., Ylanne, J.(2006) Blood 107: 1925
- PubMed: 16293600
- DOI: https://doi.org/10.1182/blood-2005-10-3964
- Primary Citation of Related Structures:
2AAV, 2BP3 - PubMed Abstract:
Filamin A (FLNa), a dimeric actin cross-linking and scaffold protein with numerous intracellular binding partners, anchors the platelet adhesion glycoprotein (GP) Ib-IX-V receptor to actin cytoskeleton. We mapped the GPIbalpha binding site to a single domain of FLNa and resolved the structure of this domain and its interaction complex with the corresponding GPIbalpha cytoplasmic domain. This is the first atomic structure of this class of membrane glycoprotein-cytoskeleton connection. GPIbalpha binds in a groove formed between the C and D beta strands of FLNa domain 17. The interaction is strikingly similar to that between the beta7 integrin tail and a different FLNa domain, potentially defining a conserved motif for FLNa binding. Nevertheless, the structures also reveal specificity of the interfaces, which explains different regulatory mechanisms. To verify the topology of GPIb-FLNa interaction we also purified the native complex from platelets and showed that GPIb interacts with the C-terminus of FLNa, which is in accordance with our biochemical and structural data.
- Hematology Division, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: