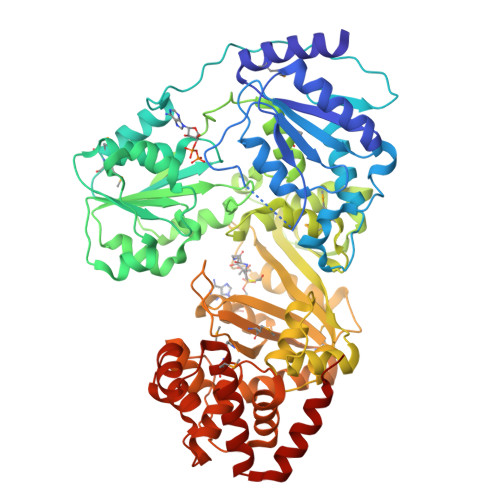
RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon
Chimnaronk, S., Suzuki, T., Manita, T., Ikeuchi, Y., Yao, M., Suzuki, T., Tanaka, I.(2009) EMBO J 28: 1362-1373
- PubMed: 19322199
- DOI: https://doi.org/10.1038/emboj.2009.69
- Primary Citation of Related Structures:
2ZPA - PubMed Abstract:
Post-transcriptional RNA modifications in the anticodon of transfer RNAs frequently contribute to the high fidelity of protein synthesis. In eubacteria, two genome-encoded transfer RNA (tRNA) species bear the same CAU sequence as the anticodons, which are differentiated by modified cytidines at the wobble positions. The elongator tRNA(Met) accepts an acetyl moiety at the wobble base to form N(4)-acetylcytidine (ac(4)C): an inherent modification ensures precise decoding of the AUG codon by strengthening C-G base-pair interaction and concurrently preventing misreading of the near cognate AUA codon. We have determined the crystal structure of tRNA(Met) cytidine acetyltransferase (TmcA) from Escherichia coli complexed with two natural ligands, acetyl-CoA and ADP, at 2.35 A resolution. The structure unexpectedly reveals an idiosyncratic RNA helicase module fused with a GCN5-related N-acetyltransferase (GNAT) fold, which intimately cross-interact. Taken together with the biochemical evidence, we further unravelled the function of acetyl-CoA as an enzyme-activating switch, and propose that an RNA helicase motor driven by ATP hydrolysis is used to deliver the wobble base to the active centre of the GNAT domain.
Organizational Affiliation:
Faculty of Advanced Life Sciences, Hokkaido University, Sapporo, Japan.