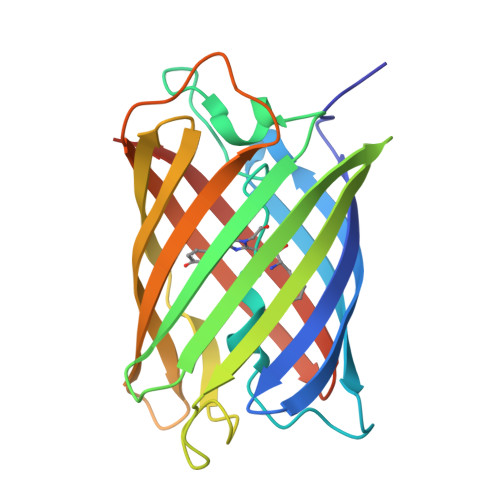
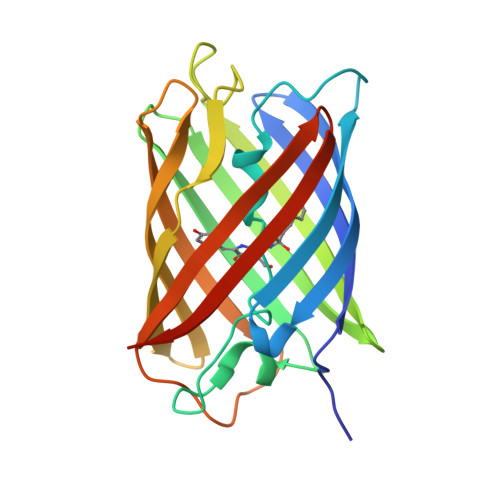
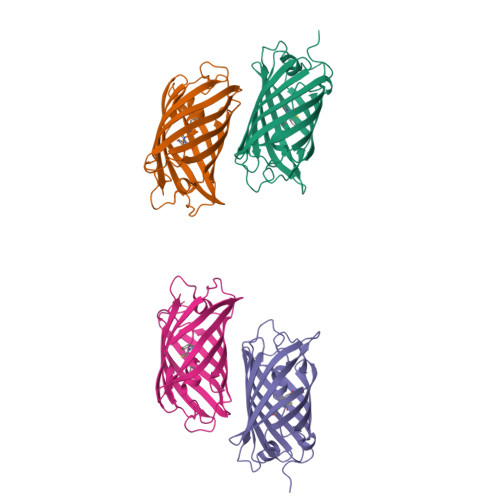Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Kikuchi, A., Fukumura, E., Karasawa, S., Mizuno, H., Miyawaki, A., Shiro, Y.(2008) Biochemistry 47: 11573-11580
- PubMed: 18844376
- DOI: https://doi.org/10.1021/bi800727v
- Primary Citation of Related Structures:
2ZMU, 2ZMW - PubMed Abstract:
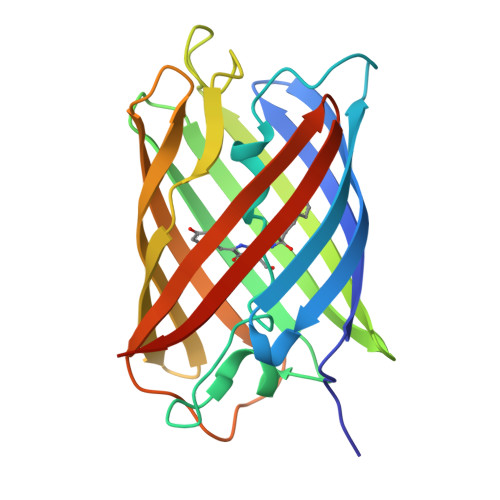
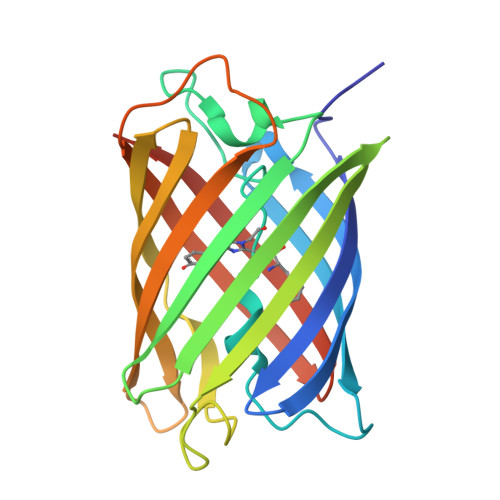
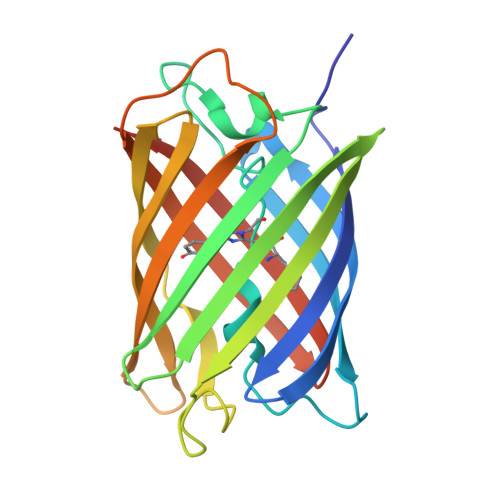
Monomeric Kusabira Orange (mKO) is a green fluorescent protein (GFP)-like protein that emits orange light at a peak of 559 nm. We analyzed its X-ray structure at 1.65 A and found a novel three-ring chromophore that developed autocatalytically from a Cys65-Tyr66-Glu67 tripeptide in which the side chain of Cys65 formed the third 2-hydroxy-3-thiazoline ring. As a result, the chromophore contained the CNCOH group at the 2-position of the imidazolinone moiety such that the conjugated pi-electron system of the chromophore was more extended than that of GFP but less extended than that of the Discosoma sp. red fluorescent protein (DsRed). Since a sulfur atom has potent nucleophilic character, the third 3-thiazoline ring is rapidly and completely cyclized. Furthermore, our structure reveals the presence of a pi-pi stacking interaction between His197 and the chromophore as well as a pi-cation interaction between Arg69 and the chromophore. These structural findings are sufficient to account for the orange emission, pH tolerance, and photostability of mKO.
Organizational Affiliation:
Biometal Science Laboratory, RIKEN SPring-8 Center, 1-1-1, Kouto, Sayo, Hyogo 679-5148, Japan. kikuchi@spring8.or.jp