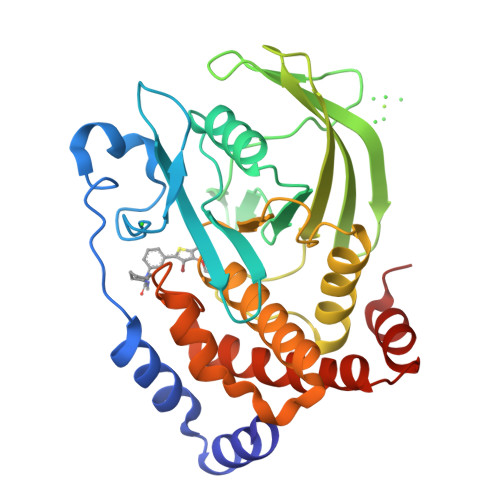
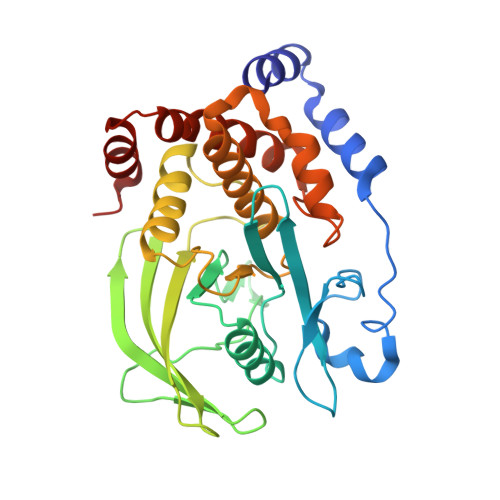
Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Wan, Z.-K., Lee, J., Hotchandani, R., Moretto, A., Binnun, E., Wilson, D.P., Kirincich, S.J., Follows, B.C., Ipek, M., Xu, W., Joseph-McCarthy, D., Zhang, Y.-L., Tam, M., Erbe, D.V., Tobin, J.F., Li, W., Tam, S.Y., Mansour, T.S., Wu, J.(2008) ChemMedChem 3: 1525-1529
- PubMed: 18798205
- DOI: https://doi.org/10.1002/cmdc.200800188
- Primary Citation of Related Structures:
2ZMM, 2ZN7
Organizational Affiliation:
Chemical and Screening Sciences, Wyeth Research, 200 Cambridgepark Drive, Cambridge, MA 02140, USA.