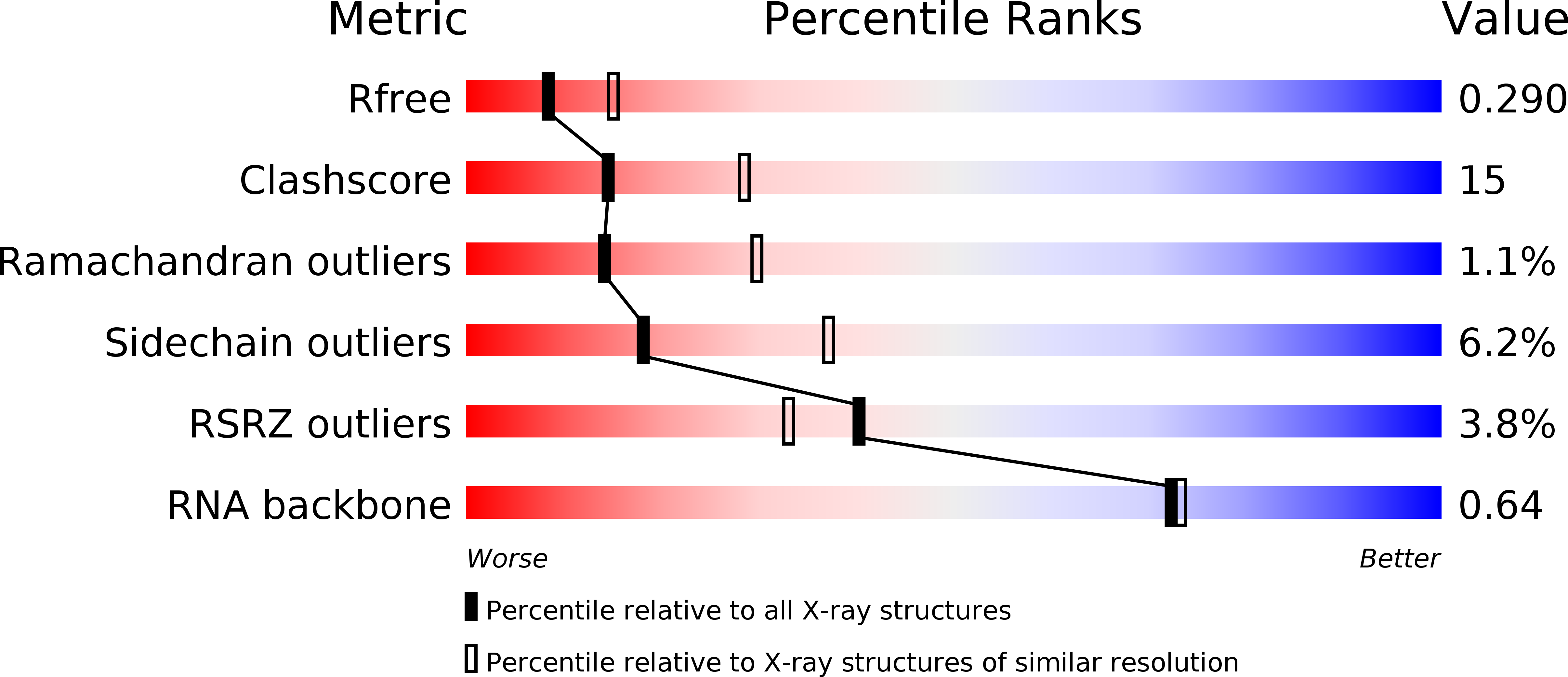
Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme
Toh, Y., Numata, T., Watanabe, K., Takeshita, D., Nureki, O., Tomita, K.(2008) EMBO J 27: 1944-1952
- PubMed: 18583961
- DOI: https://doi.org/10.1038/emboj.2008.124
- Primary Citation of Related Structures:
2ZH1, 2ZH2, 2ZH3, 2ZH4, 2ZH5, 2ZH6, 2ZH7, 2ZH8, 2ZH9, 2ZHA, 2ZHB - PubMed Abstract:
CCA-adding enzyme builds the 3'-end CCA of tRNA without a nucleic acid template. The mechanism for the maintenance of fidelity during the CCA-adding reaction remains elusive. Here, we present almost a dozen complex structures of the class I CCA-adding enzyme and tRNA mini-helices (mini-D(73)N(74), mini-D(73)N(74)C(75) and mini-D(73)C(74)N(75); D(73) is a discriminator nucleotide and N is either A, G, or U). The mini-D(73)N(74) complexes adopt catalytically inactive open forms, and CTP shifts the enzymes to the active closed forms and allows N(74) to flip for CMP incorporation. In contrast, unlike the catalytically active closed form of the mini-D(73)C(74)C(75) complex, the mini-D(73)N(74)C(75) and mini-D(73)C(74)N(75) complexes adopt inactive open forms. Only the mini-D(73)C(74)U(75) accepts AMP to a similar extent as mini-D(73)C(74)C(75), and ATP shifts the enzyme to a closed, active form and allows U(75) to flip for AMP incorporation. These findings suggest that the 3'-region of RNA is proofread, after two nucleotide additions, in the closed, active form of the complex at the AMP incorporation stage. This proofreading is a prerequisite for the maintenance of fidelity for complete CCA synthesis.
Organizational Affiliation:
Institute of Biological Resources and Functions, National Institute of Advanced Industrial Science and Technology (AIST), Ibaraki, Japan.