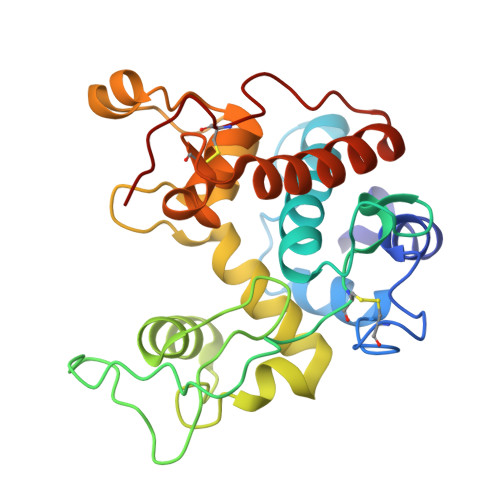
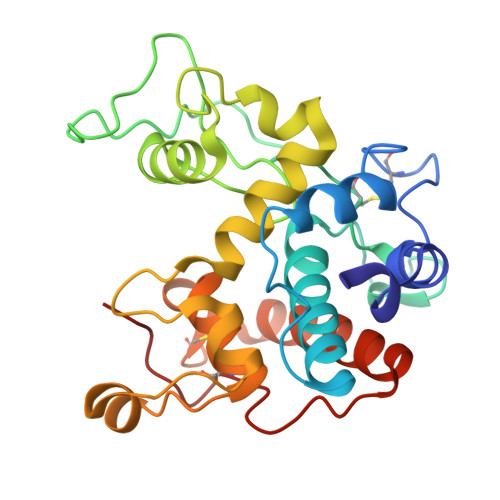
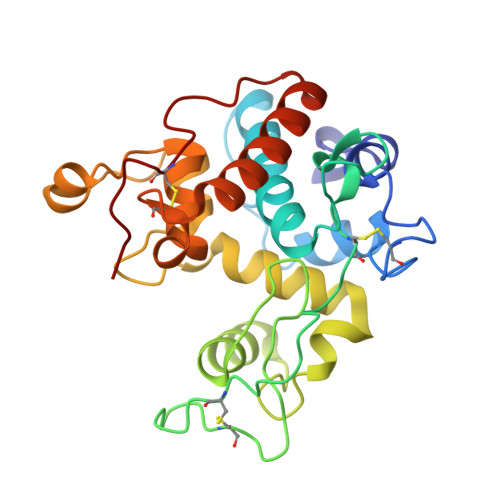
Crystal structures of a family 19 chitinase from Brassica juncea show flexibility of binding cleft loops
Ubhayasekera, W., Tang, C.M., Ho, S.W.T., Berglund, G., Bergfors, T., Chye, M.-L., Mowbray, S.L.(2007) FEBS J 274: 3695-3703
- PubMed: 17608716
- DOI: https://doi.org/10.1111/j.1742-4658.2007.05906.x
- Primary Citation of Related Structures:
2Z37, 2Z38, 2Z39 - PubMed Abstract:
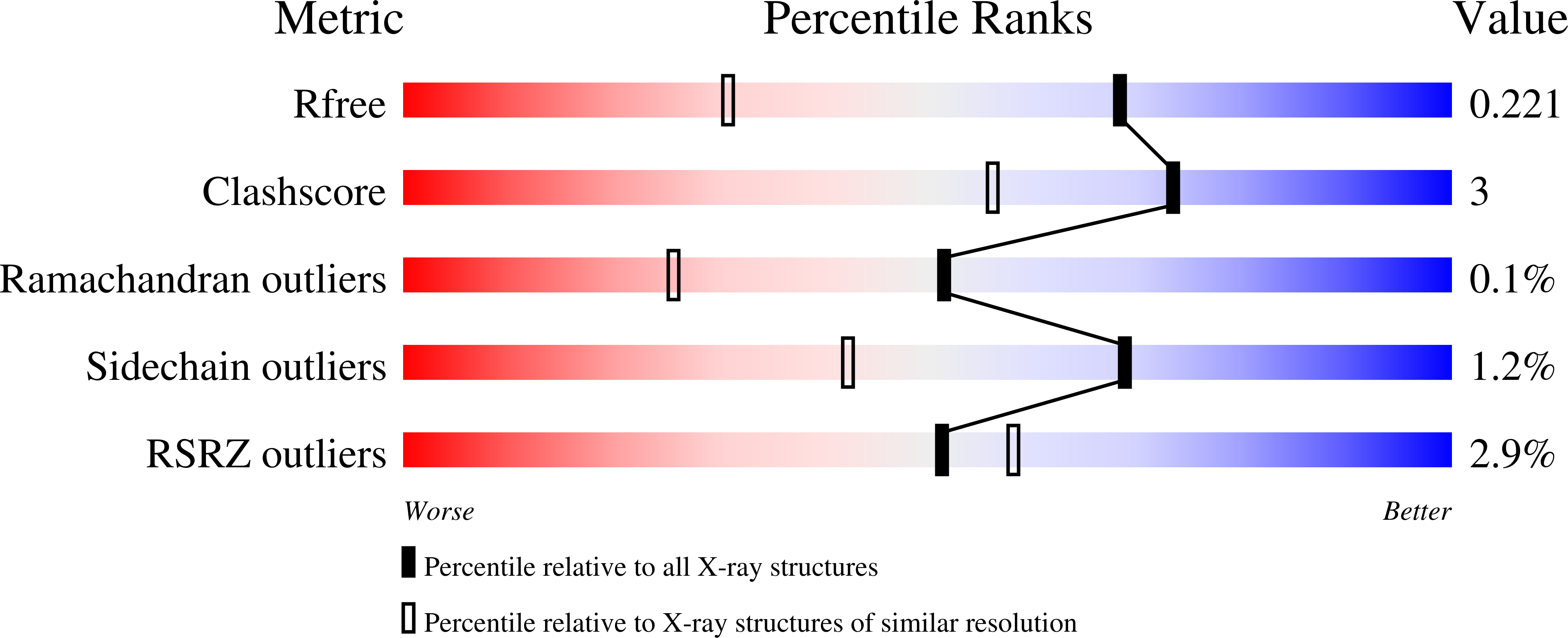
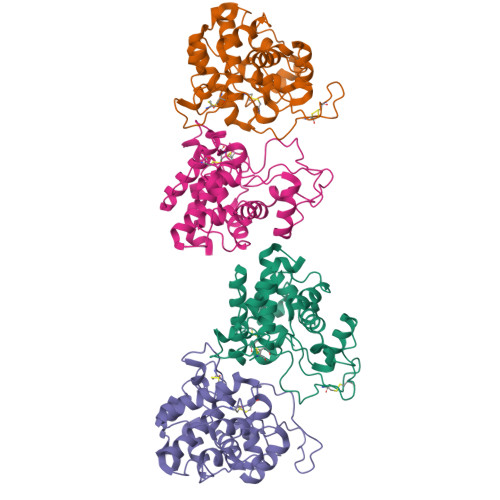
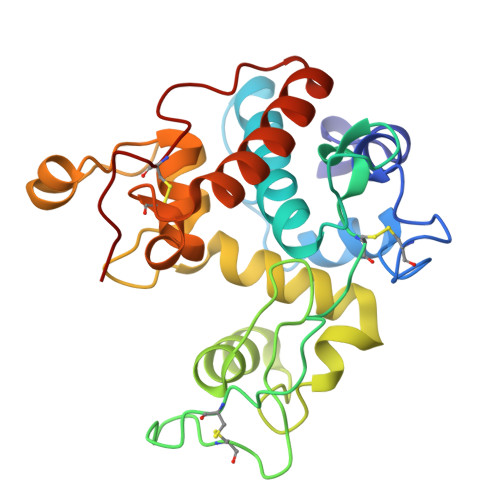
Brassica juncea chitinase is an endo-acting, pathogenesis-related protein that is classified into glycoside hydrolase family 19, with highest homology (50-60%) in its catalytic domain to class I plant chitinases. Here we report X-ray structures of the chitinase catalytic domain from wild-type (apo, as well as with chloride ions bound) and a Glu234Ala mutant enzyme, solved by molecular replacement and refined at 1.53, 1.8 and 1.7 A resolution, respectively. Confirming our earlier mutagenesis studies, the active-site residues are identified as Glu212 and Glu234. Glu212 is believed to be the catalytic acid in the reaction, whereas Glu234 is thought to have a dual role, both activating a water molecule in its attack on the anomeric carbon, and stabilizing the charged intermediate. The molecules in the various structures differ significantly in the conformation of a number of loops that border the active-site cleft. The differences suggest an opening and closing of the enzyme during the catalytic cycle. Chitin is expected to dock first near Glu212, which will protonate it. Conformational changes then bring Glu234 closer, allowing it to assist in the following steps. These observations provide important insights into catalysis in family 19 chitinases.
Organizational Affiliation:
Department of Molecular Biology, Swedish University of Agricultural Sciences, Uppsala, Sweden Department of Botany, The University of Hong Kong, Pokfulam, Hong Kong Department of Cell and Molecular Biology, Uppsala University, Sweden.