Structural Basis for Substrate Recognition by Erwinia Chrysanthemi Gh30 Glucuronoxylanase.
Urbanikova, L., Vrsanska, M., Krogh, K.B., Hoff, T., Biely, P.(2011) FEBS J 278: 2105
- PubMed: 21501386
- DOI: https://doi.org/10.1111/j.1742-4658.2011.08127.x
- Primary Citation of Related Structures:
2Y24 - PubMed Abstract:
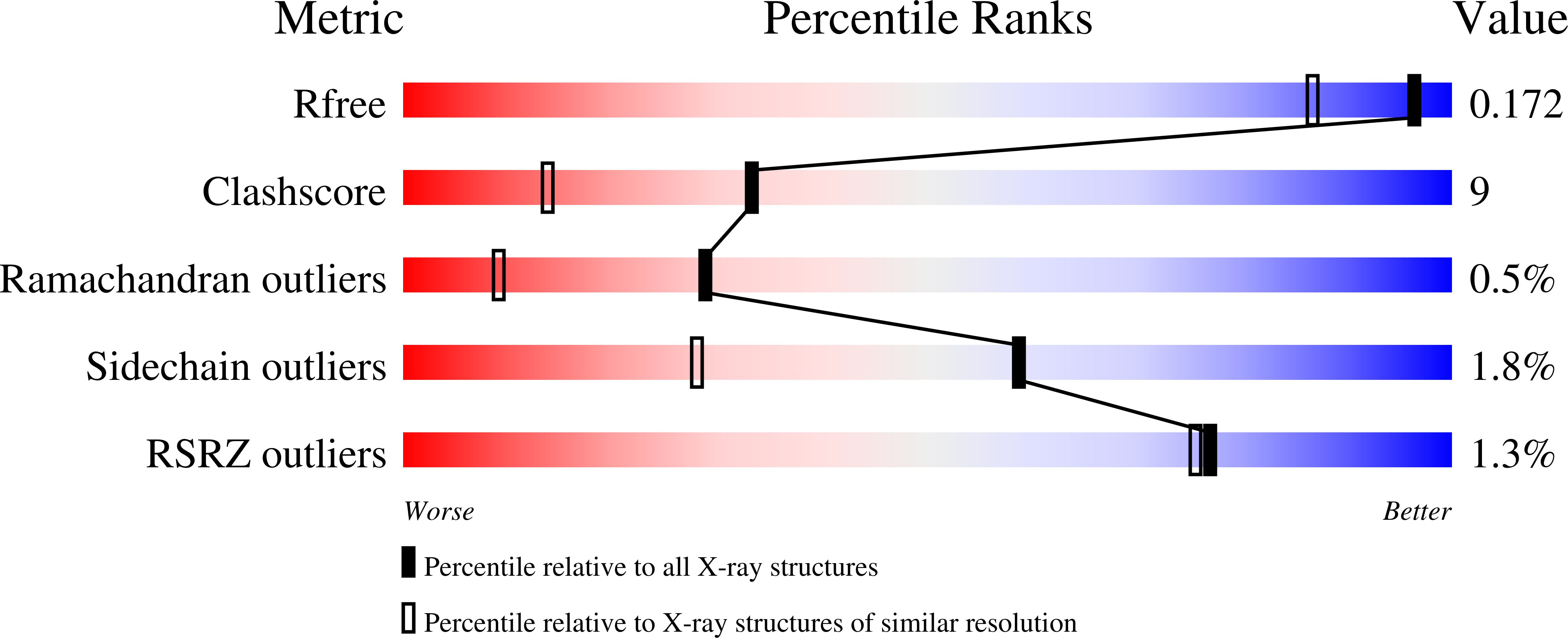
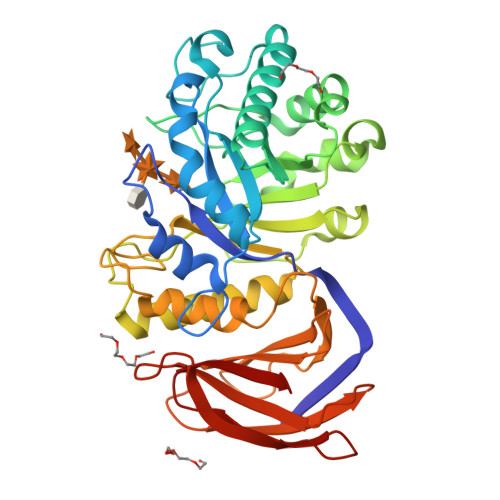
Xylanase A from the phytopathogenic bacterium Erwinia chrysanthemi is classified as a glycoside hydrolase family 30 enzyme (previously in family 5) and is specialized for degradation of glucuronoxylan. The recombinant enzyme was crystallized with the aldotetraouronic acid β-D-xylopyranosyl-(1→4)-[4-O-methyl-α-D-glucuronosyl-(1→2)]-β-D-xylopyranosyl-(1→4)-D-xylose as a ligand. The crystal structure of the enzyme-ligand complex was solved at 1.39 Å resolution. The ligand xylotriose moiety occupies subsites -1, -2 and -3, whereas the methyl glucuronic acid residue attached to the middle xylopyranosyl residue of xylotriose is bound to the enzyme through hydrogen bonds to five amino acids and by the ionic interaction of the methyl glucuronic acid carboxylate with the positively charged guanidinium group of Arg293. The interaction of the enzyme with the methyl glucuronic acid residue appears to be indispensable for proper distortion of the xylan chain and its effective hydrolysis. Such a distortion does not occur with linear β-1,4-xylooligosaccharides, which are hydrolyzed by the enzyme at a negligible rate. Structural and experimental data are available in the Protein Data Bank database under accession number 2y24 [45].
Organizational Affiliation:
Institute of Molecular Biology, Slovak Academy of Sciences, Bratislava, Slovakia.