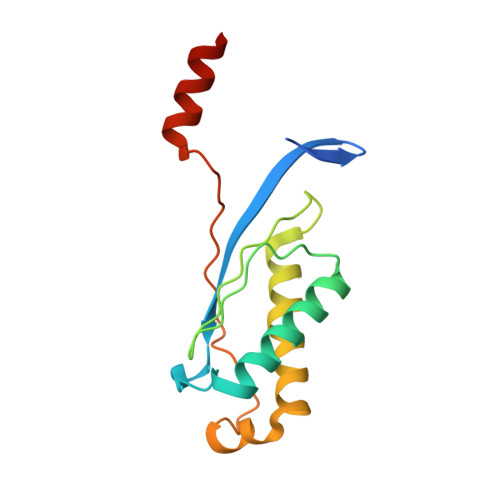
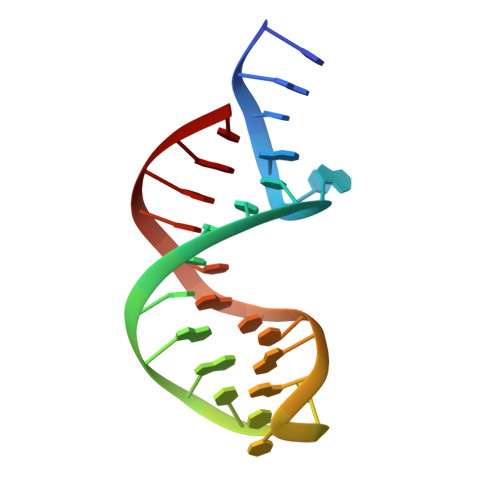
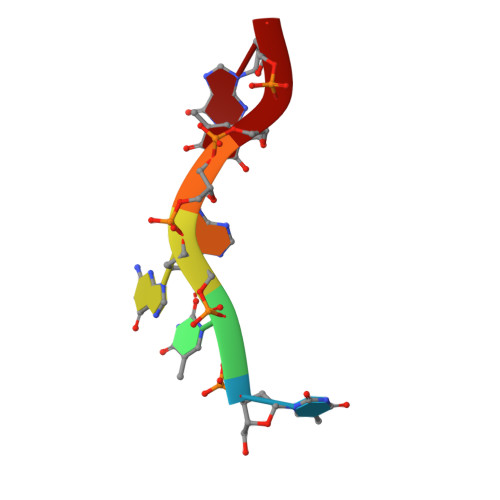
DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Hickman, A.B., James, J.A., Barabas, O., Pasternak, C., Ton-Hoang, B., Chandler, M., Sommer, S., Dyda, F.(2010) EMBO J 29: 3840
- PubMed: 20890269
- DOI: https://doi.org/10.1038/emboj.2010.241
- Primary Citation of Related Structures:
2XM3, 2XMA, 2XO6, 2XQC - PubMed Abstract:
Bacterial insertion sequences (ISs) from the IS200/IS605 family encode the smallest known DNA transposases and mobilize through single-stranded DNA transposition. Transposition by one particular family member, ISDra2 from Deinococcus radiodurans, is dramatically stimulated upon massive γ irradiation. We have determined the crystal structures of four ISDra2 transposase/IS end complexes; combined with in vivo activity assays and fluorescence anisotropy binding measurements, these have revealed the molecular basis of strand discrimination and transposase action. The structures also show that previously established structural rules of target site recognition that allow different specific sequences to be targeted are only partially conserved among family members. Furthermore, we have captured a fully assembled active site including the scissile phosphate bound by a divalent metal ion cofactor (Cd²(+)) that supports DNA cleavage. Finally, the observed active site rearrangements when the transposase binds a metal ion in which it is inactive provide a clear rationale for metal ion specificity.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes, Digestive, and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.