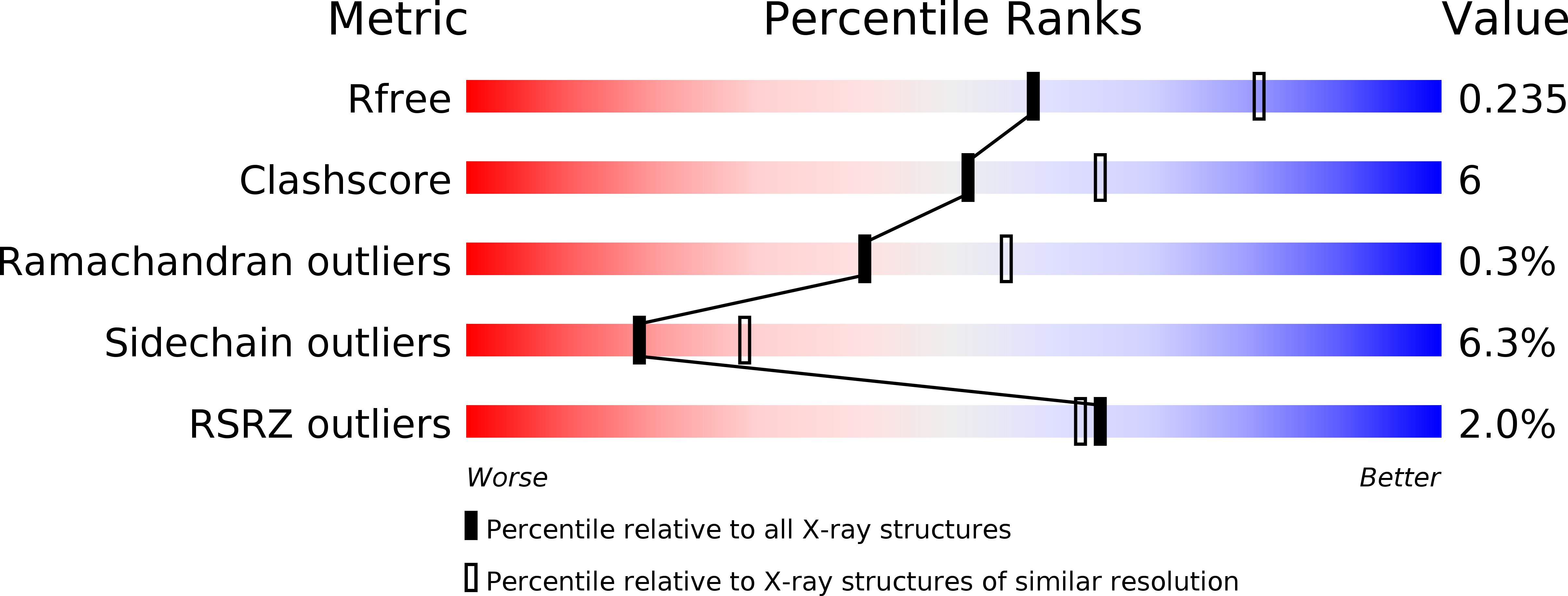
Cell-Penetrant, Nanomolar O-Glcnacase Inhibitors Selective Against Lysosomal Hexosaminidases.
Dorfmueller, H.C., Borodkin, V.S., Schimpl, M., Zheng, X., Kime, R., Read, K.D., Van Aalten, D.M.F.(2010) Chem Biol 17: 1250
- PubMed: 21095575
- DOI: https://doi.org/10.1016/j.chembiol.2010.09.014
- Primary Citation of Related Structures:
2XPK - PubMed Abstract:
Posttranslational modification of metazoan nucleocytoplasmic proteins with N-acetylglucosamine (O-GlcNAc) is essential, dynamic, and inducible and can compete with protein phosphorylation in signal transduction. Inhibitors of O-GlcNAcase, the enzyme removing O-GlcNAc, are useful tools for studying the role of O-GlcNAc in a range of cellular processes. We report the discovery of nanomolar OGA inhibitors that are up to 900,000-fold selective over the related lysosomal hexosaminidases. When applied at nanomolar concentrations on live cells, these cell-penetrant molecules shift the O-GlcNAc equilibrium toward hyper-O-GlcNAcylation with EC₅₀ values down to 3 nM and are thus invaluable tools for the study of O-GlcNAc cell biology.
Organizational Affiliation:
Division of Molecular Microbiology, College of Life Sciences, University of Dundee, Dundee DD15EH, Scotland.