Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria.
Watt, I.N., Montgomery, M.G., Runswick, M.J., Leslie, A.G.W., Walker, J.E.(2010) Proc Natl Acad Sci U S A 107: 16823
- PubMed: 20847295
- DOI: https://doi.org/10.1073/pnas.1011099107
- Primary Citation of Related Structures:
2XND - PubMed Abstract:
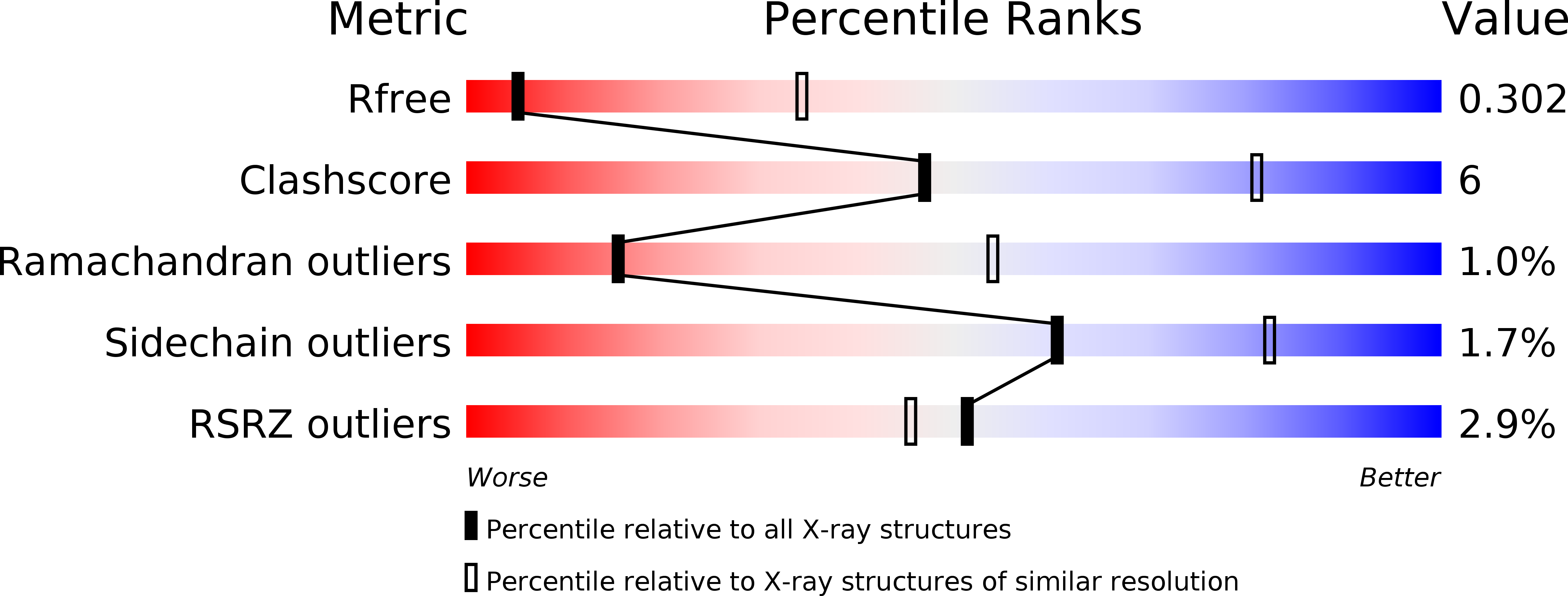
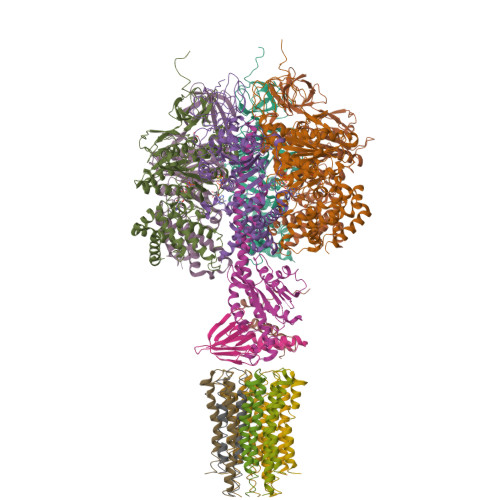
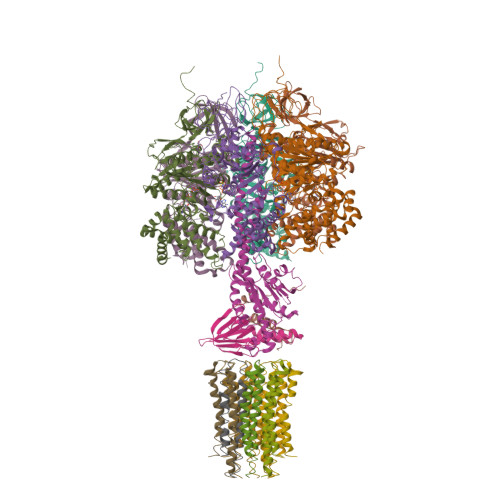
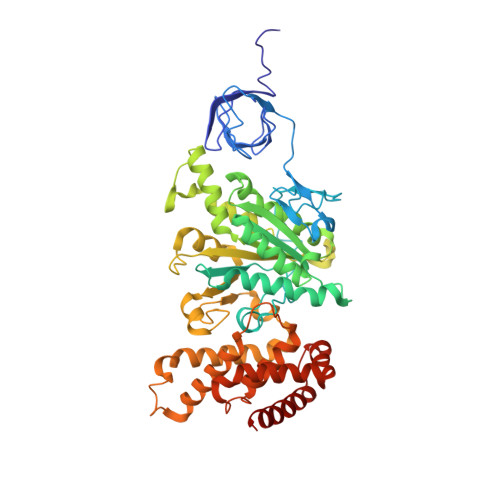
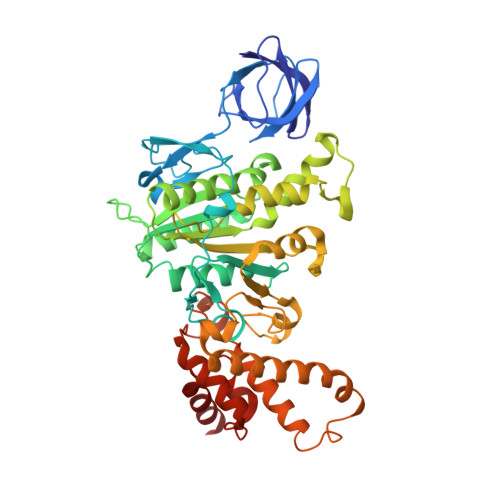
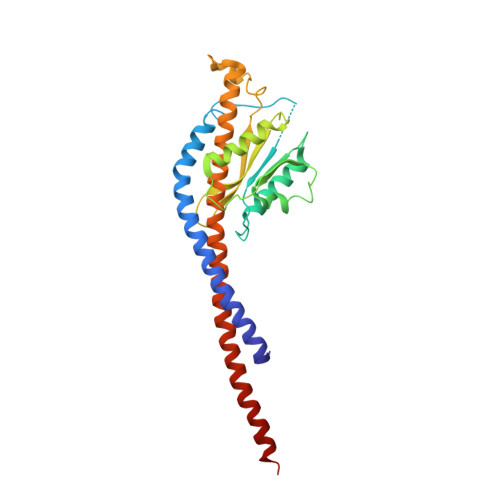
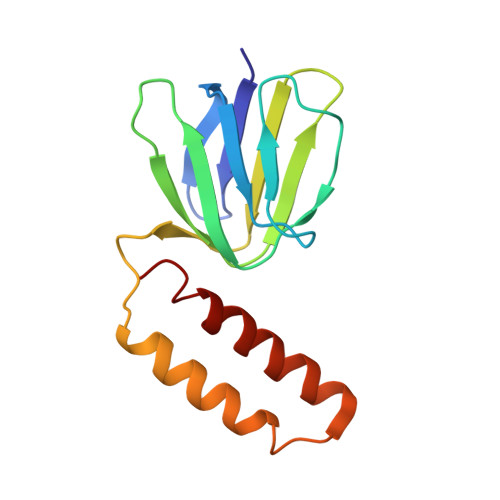
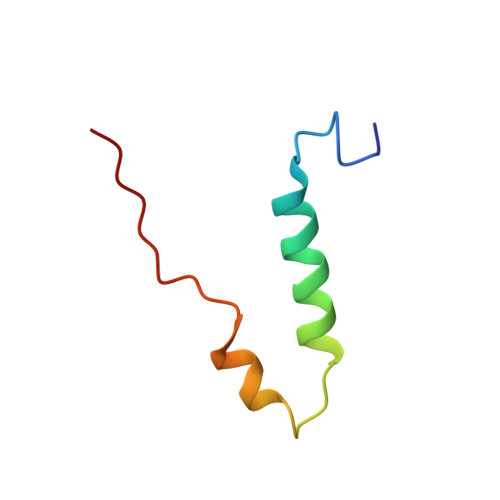
The catalytic domain of the F-ATPase in mitochondria protrudes into the matrix of the organelle, and is attached to the membrane domain by central and peripheral stalks. Energy for the synthesis of ATP from ADP and phosphate is provided by the transmembrane proton-motive-force across the inner membrane, generated by respiration. The proton-motive force is coupled mechanically to ATP synthesis by the rotation at about 100 times per second of the central stalk and an attached ring of c-subunits in the membrane domain. Each c-subunit carries a glutamate exposed around the midpoint of the membrane on the external surface of the ring. The rotation is generated by protonation and deprotonation successively of each glutamate. Each 360° rotation produces three ATP molecules, and requires the translocation of one proton per glutamate by each c-subunit in the ring. In fungi, eubacteria, and plant chloroplasts, ring sizes of c(10)-c(15) subunits have been observed, implying that these enzymes need 3.3-5 protons to make each ATP, but until now no higher eukaryote has been examined. As shown here in the structure of the bovine F(1)-c-ring complex, the c-ring has eight c-subunits. As the sequences of c-subunits are identical throughout almost all vertebrates and are highly conserved in invertebrates, their F-ATPases probably contain c(8)-rings also. Therefore, in about 50,000 vertebrate species, and probably in many or all of the two million invertebrate species, 2.7 protons are required by the F-ATPase to make each ATP molecule.
Organizational Affiliation:
The Medical Research Council Mitochondrial Biology Unit, Hills Road, Cambridge, CB2 0XY, UK.