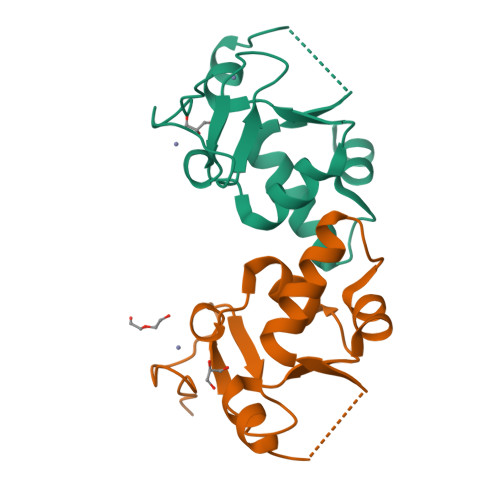
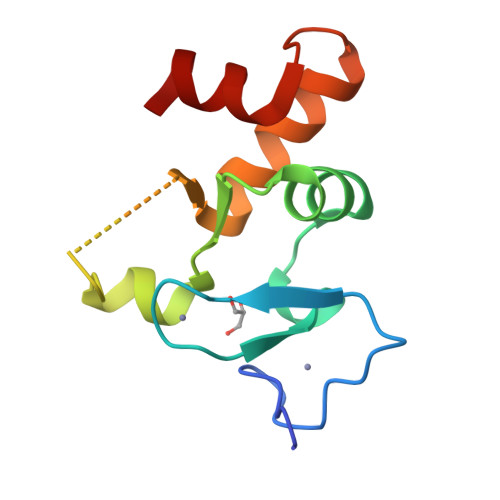
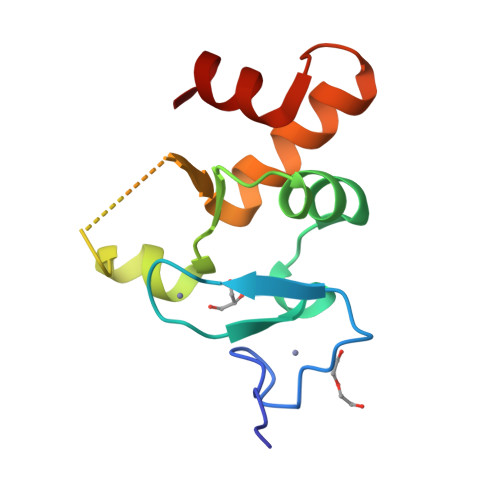
Allosteric Remodeling of the Histone H3 Binding Pocket in the Pygo2 Phd Finger Triggered by its Binding to the B9L/Bcl9 Co-Factor.
Miller, T.C., Rutherford, T.J., Johnson, C.M., Fiedler, M., Bienz, M.(2010) J Mol Biology 401: 969
- PubMed: 20637214
- DOI: https://doi.org/10.1016/j.jmb.2010.07.007
- Primary Citation of Related Structures:
2XB1 - PubMed Abstract:
The Zn-coordinated PHD fingers of Pygopus (Pygo) proteins are critical for beta-catenin-dependent transcriptional switches in normal and malignant tissues. They bind to methylated histone H3 tails, assisted by their BCL9 co-factors whose homology domain 1 (HD1) binds to the rear PHD surface. Although histone-binding residues are identical between the two human Pygo paralogs, we show here that Pygo2 complexes exhibit slightly higher binding affinities for methylated histone H3 tail peptides than Pygo1 complexes. We solved the crystal structure of the Pygo2 PHD-BCL9-2 HD1 complex, which revealed paralog-specific interactions in its PHD-HD1 interface that could contribute indirectly to its elevated affinity for the methylated histone H3 tail. Interestingly, using NMR spectroscopy, we discovered that HD1 binding to PHD triggers an allosteric communication with a conserved isoleucine residue that lines the binding channel for histone H3 threonine 3 (T3), the link between the two adjacent binding pockets accommodating histone H3 alanine 1 and methylated lysine 4, respectively. This modulates the surface of the T3 channel, providing a plausible explanation as to how BCL9 co-factors binding to Pygo PHD fingers impact indirectly on their histone binding affinity. Intriguingly, this allosteric modulation of the T3 channel is propagated through the PHD structural core by a highly conserved tryptophan, the signature residue defining the PHD subclass of Zn fingers, which suggests that other PHD proteins may also be assisted by co-factors in their decoding of modified histone H3 tails.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.