The Conserved Candida Albicans Ca3427 Gene Product Defines a New Family of Proteins Exhibiting the Generic Periplasmic Binding Protein Structural Fold
Santini, S., Claverie, J.M., Mouz, N., Rousselle, T., Maza, C., Monchois, V., Abergel, C.(2011) PLoS One 6: 18528
- PubMed: 21494601
- DOI: https://doi.org/10.1371/journal.pone.0018528
- Primary Citation of Related Structures:
2X7P, 2X7Q - PubMed Abstract:
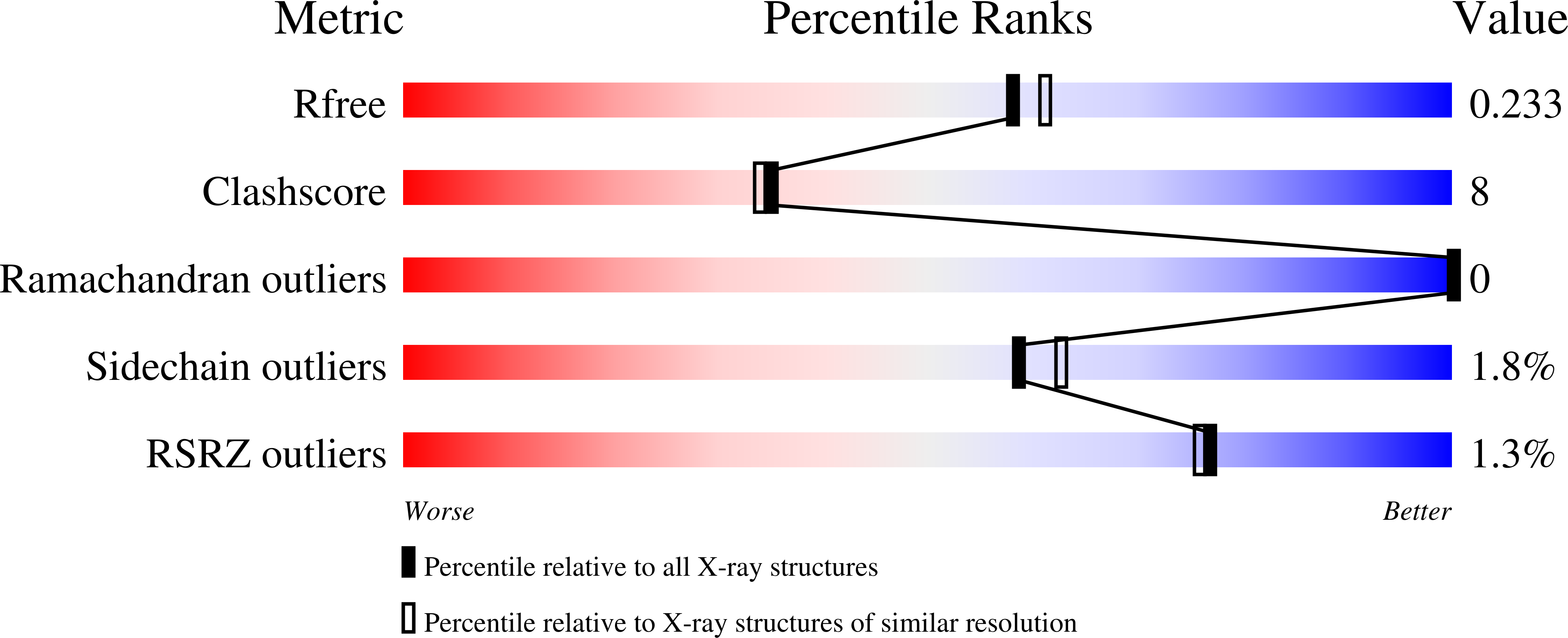
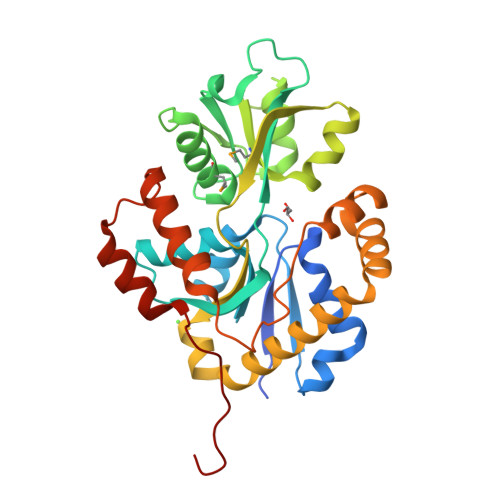
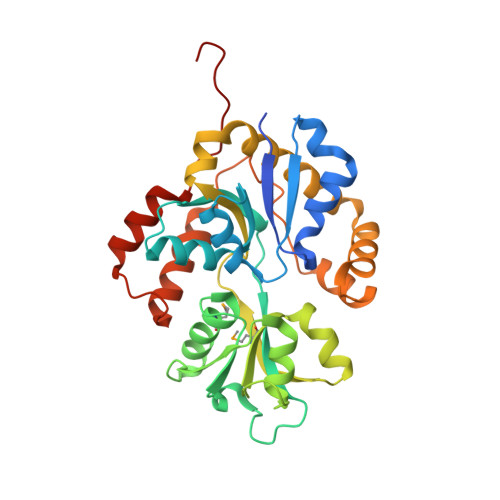
Nosocomial diseases due to Candida albicans infections are in constant rise in hospitals, where they cause serious complications to already fragile intensive care patients. Antifungal drug resistance is fast becoming a serious issue due to the emergence of strains resistant to currently available antifungal agents. Thus the urgency to identify new potential protein targets, the function and structure of which may guide the development of new antifungal drugs. In this context, we initiated a comparative genomics study in search of promising protein coding genes among the most conserved ones in reference fungal genomes. The CA3427 gene was selected on the basis of its presence among pathogenic fungi contrasting with its absence in the non pathogenic Saccharomyces cerevisiae. We report the crystal 3D-structure of the Candida albicans CA3427 protein at 2.1 Å resolution. The combined analysis of its sequence and structure reveals a structural fold originally associated with periplasmic binding proteins. The CA3427 structure highlights a binding site located between the two protein domains, corresponding to a sequence segment conserved among fungi. Two crystal forms of CA3427 were found, suggesting that the presence or absence of a ligand at the proposed binding site might trigger a "Venus flytrap" motion, coupled to the previously described activity of bacterial periplasmic binding proteins. The conserved binding site defines a new subfamily of periplasmic binding proteins also found in many bacteria of the bacteroidetes division, in a choanoflagellate (a free-living unicellular and colonial flagellate eukaryote) and in a placozoan (the closest multicellular relative of animals). A phylogenetic analysis suggests that this gene family originated in bacteria before its horizontal transfer to an ancestral eukaryote prior to the radiation of fungi. It was then lost by the Saccharomycetales which include Saccharomyces cerevisiae.
Organizational Affiliation:
Information Génomique et Structurale (CNRS UPR2589), Aix-Marseille Université, Mediterranean Institute of Microbiology, Parc Scientifique de Luminy, Marseille, France.