Polymerization of Mip-1 Chemokine (Ccl3 and Ccl4) and Clearance of Mip-1 by Insulin-Degrading Enzyme.
Ren, M., Guo, Q., Guo, L., Lenz, M., Qian, F., Koenen, R.R., Xu, H., Schilling, A.B., Weber, C., Ye, R.D., Dinner, A.R., Tang, W.(2010) EMBO J 29: 3952
- PubMed: 20959807
- DOI: https://doi.org/10.1038/emboj.2010.256
- Primary Citation of Related Structures:
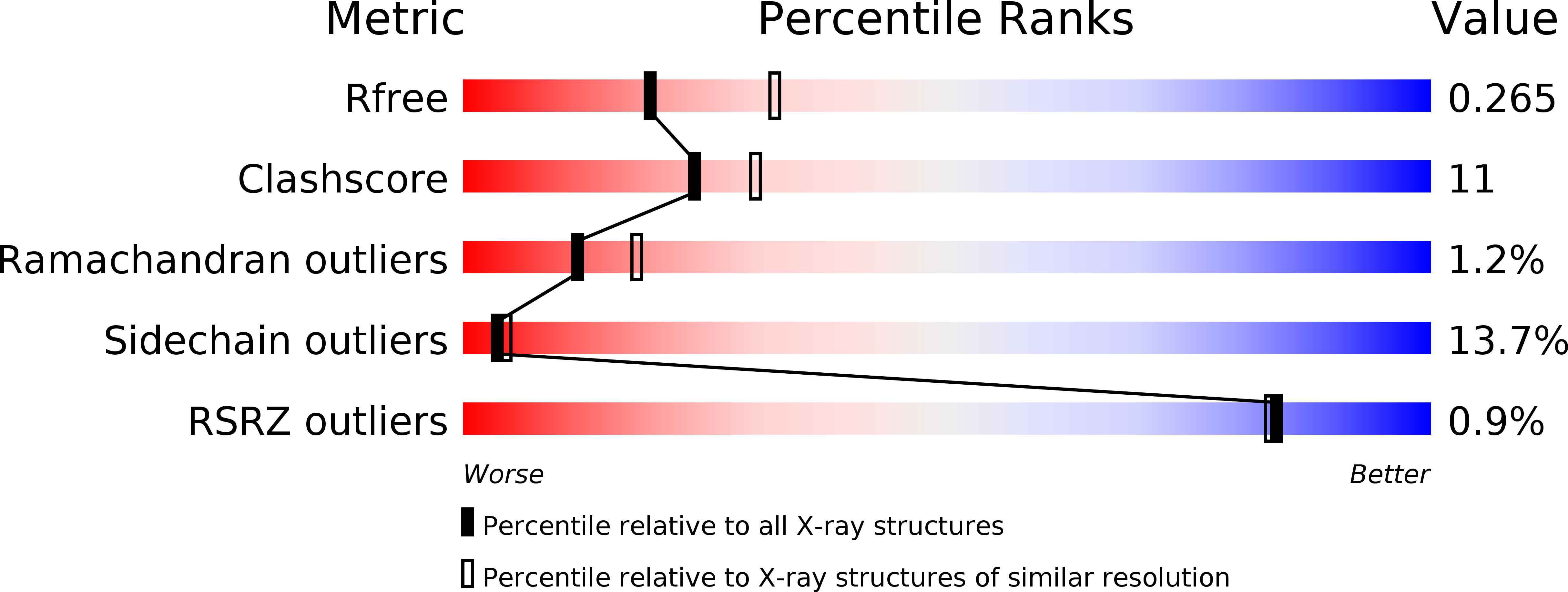
2X69, 2X6G, 2X6L - PubMed Abstract:
Macrophage inflammatory protein-1 (MIP-1), MIP-1α (CCL3) and MIP-1β (CCL4) are chemokines crucial for immune responses towards infection and inflammation. Both MIP-1α and MIP-1β form high-molecular-weight aggregates. Our crystal structures reveal that MIP-1 aggregation is a polymerization process and human MIP-1α and MIP-1β form rod-shaped, double-helical polymers. Biophysical analyses and mathematical modelling show that MIP-1 reversibly forms a polydisperse distribution of rod-shaped polymers in solution. Polymerization buries receptor-binding sites of MIP-1α, thus depolymerization mutations enhance MIP-1α to arrest monocytes onto activated human endothelium. However, same depolymerization mutations render MIP-1α ineffective in mouse peritoneal cell recruitment. Mathematical modelling reveals that, for a long-range chemotaxis of MIP-1, polymerization could protect MIP-1 from proteases that selectively degrade monomeric MIP-1. Insulin-degrading enzyme (IDE) is identified as such a protease and decreased expression of IDE leads to elevated MIP-1 levels in microglial cells. Our structural and proteomic studies offer a molecular basis for selective degradation of MIP-1. The regulated MIP-1 polymerization and selective inactivation of MIP-1 monomers by IDE could aid in controlling the MIP-1 chemotactic gradient for immune surveillance.
Organizational Affiliation:
Ben-May Department for Cancer Research, The University of Chicago, Chicago, IL 60637, USA.