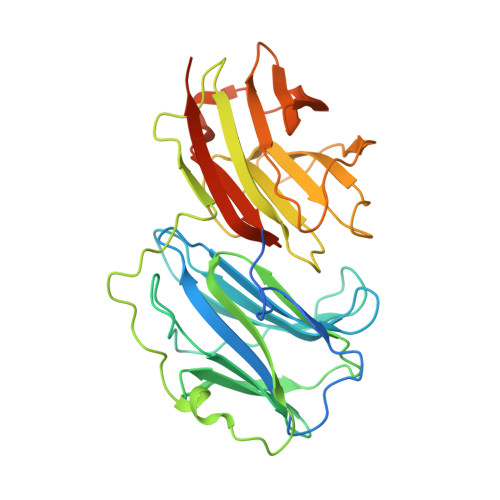
Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains.
Guardado-Calvo, P., Munoz, E.M., Llamas-Saiz, A.L., Fox, G.C., Kahn, R., Curiel, D.T., Glasgow, J.N., van Raaij, M.J.(2010) J Virol 84: 10558-10568
- PubMed: 20686025
- DOI: https://doi.org/10.1128/JVI.00997-10
- Primary Citation of Related Structures:
2WST, 2WSU, 2WSV, 2WT0, 2WT1, 2WT2 - PubMed Abstract:
Adenovirus isolate NADC-1, a strain of porcine adenovirus type 4, has a fiber containing an N-terminal virus attachment region, shaft and head domains, and a C-terminal galectin domain connected to the head by an RGD-containing sequence. The crystal structure of the head domain is similar to previously solved adenovirus fiber head domains, but specific residues for binding the coxsackievirus and adenovirus receptor (CAR), CD46, or sialic acid are not conserved. The structure of the galectin domain reveals an interaction interface between its two carbohydrate recognition domains, locating both sugar binding sites face to face. Sequence evidence suggests other tandem-repeat galectins have the same arrangement. We show that the galectin domain binds carbohydrates containing lactose and N-acetyl-lactosamine units, and we present structures of the galectin domain with lactose, N-acetyl-lactosamine, 3-aminopropyl-lacto-N-neotetraose, and 2-aminoethyl-tri(N-acetyl-lactosamine), confirming the domain as a bona fide galectin domain.
Organizational Affiliation:
Departamento de Bioquímica y Biología Molecular, Facultad de Farmacia, Universidad de Santiago de Compostela, E-15782 Santiago de Compostela, Spain.