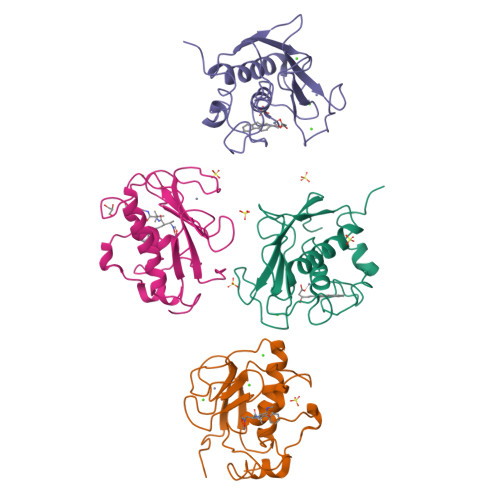
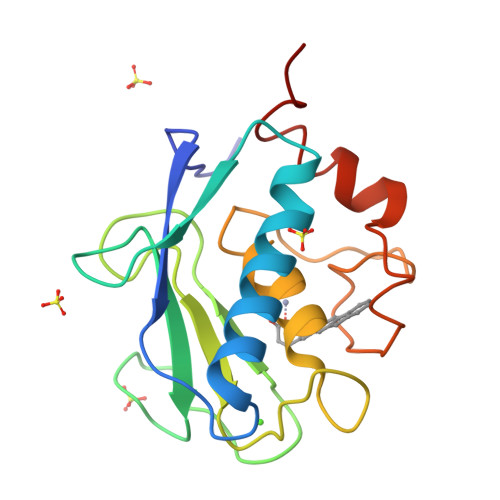
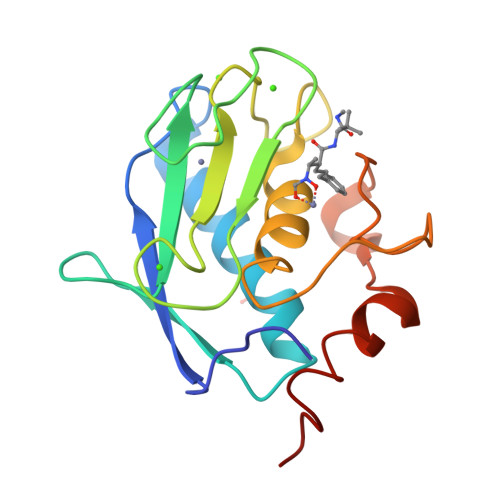
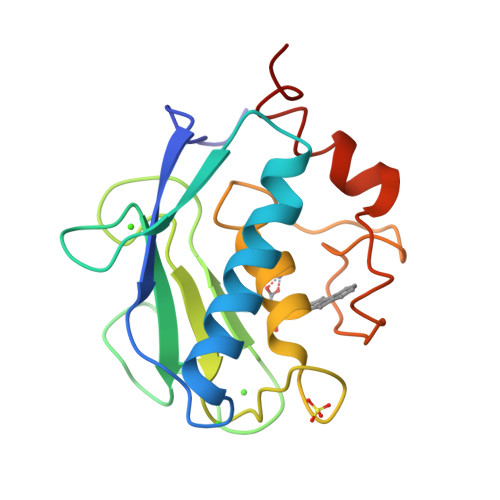
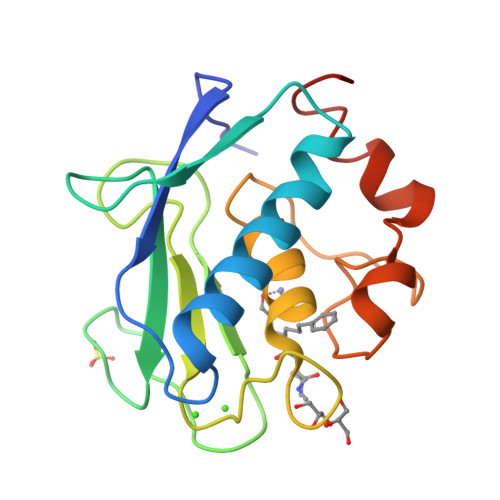
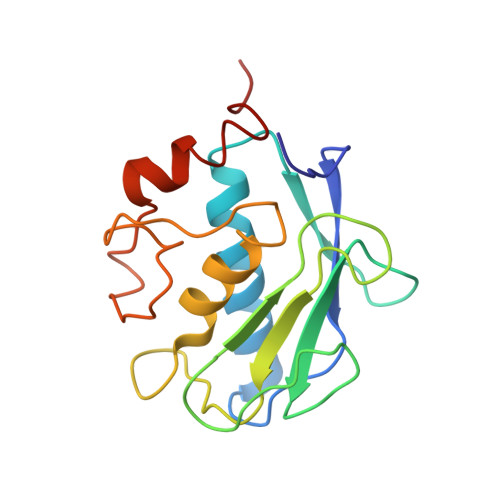
The Identification of Beta-Hydroxy Carboxylic Acids as Selective Mmp-12 Inhibitors.
Holmes, I.P., Gaines, S., Watson, S.P., Lorthioir, O., Walker, A., Baddeley, S.J., Herbert, S., Egan, D., Convery, M.A., Singh, O.M.P., Gross, J.W., Strelow, J.M., Smith, R.H., Amour, A.J., Brown, D., Martin, S.L.(2009) Bioorg Med Chem Lett 19: 5760
- PubMed: 19703773
- DOI: https://doi.org/10.1016/j.bmcl.2009.07.155
- Primary Citation of Related Structures:
2WO8, 2WO9, 2WOA - PubMed Abstract:
A new class of selective MMP-12 inhibitors have been identified via high throughput screening. Crystallization with MMP-12 confirmed the mode of binding and allowed initial optimization to be carried out using classical structure based design.
Organizational Affiliation:
GSK Medicines Research Centre, Gunnels Wood Road, Stevenage, Hertfordshire SG1 2NY, United Kingdom.