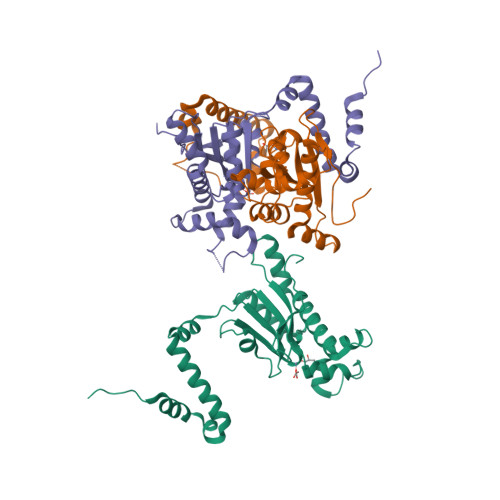
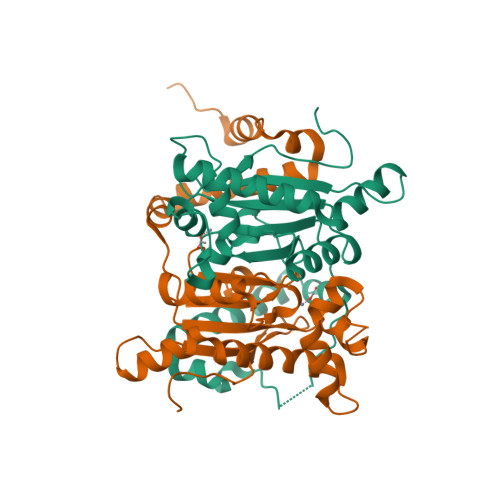
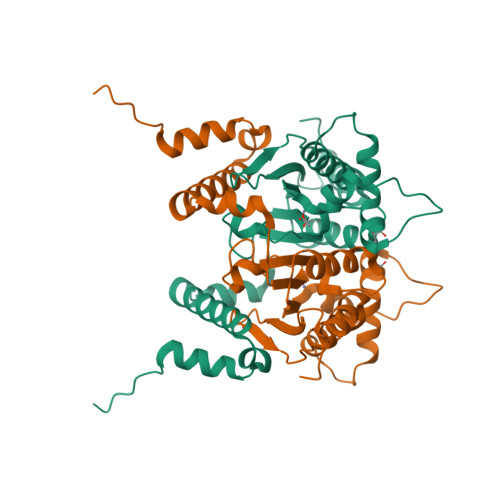
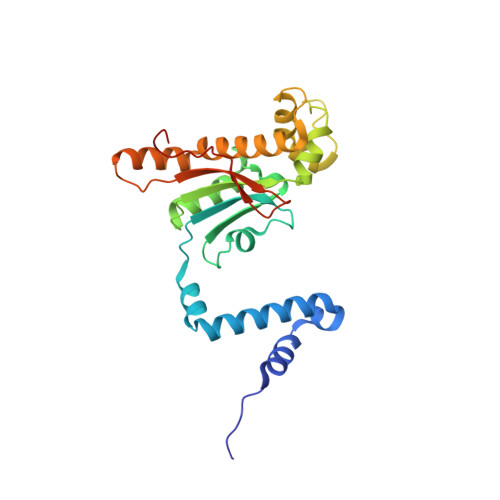
Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans.
Schlicker, C., Hall, R.A., Vullo, D., Middelhaufe, S., Gertz, M., Supuran, C.T., Muhlschlegel, F.A., Steegborn, C.(2009) J Mol Biology 385: 1207-1220
- PubMed: 19071134
- DOI: https://doi.org/10.1016/j.jmb.2008.11.037
- Primary Citation of Related Structures:
2W3N, 2W3Q - PubMed Abstract:
In the pathogenic fungus Cryptococcus neoformans, a CO(2)-sensing system is essential for survival in the natural environment (approximately 0.03% CO(2)) and mediates the switch to virulent growth in the human host (approximately 5% CO(2)). This system is composed of the carbonic anhydrase (CA) Can2, which catalyzes formation of bicarbonate, and the fungal, bicarbonate-stimulated adenylyl cyclase Cac1. The critical role of these enzymes for fungal metabolism and pathogenesis identifies them as targets for antifungal drugs. Here, we prove functional similarity of Can2 to the CA Nce103 from Candida albicans and describe its biochemical and structural characterization. The crystal structure of Can2 reveals that the enzyme belongs to the "plant-type" beta-CAs but carries a unique N-terminal extension that can interact with the active-site entrance of the dimer. We further tested a panel of compounds, identifying nanomolar Can2 inhibitors, and present the structure of a Can2 complex with the inhibitor and product analog acetate, revealing insights into interactions with physiological ligands and inhibitors.
- Department of Physiological Chemistry, Ruhr-University Bochum, Universitätsstrasse 150, 44801 Bochum, Germany.
Organizational Affiliation: