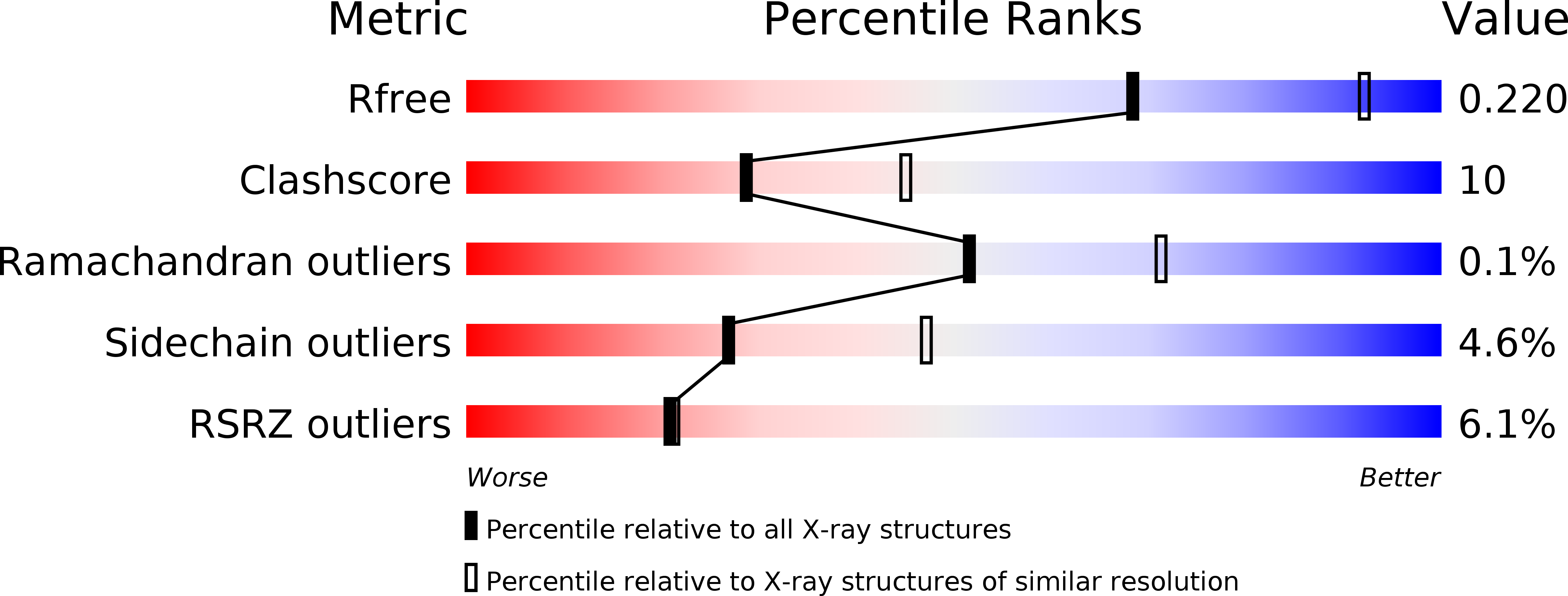
Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Pirrat, P., Smith, M.A., Pearson, A.R., McPherson, M.J., Phillips, S.E.V.(2008) Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 1105
- PubMed: 19052360
- DOI: https://doi.org/10.1107/S1744309108036373
- Primary Citation of Related Structures:
2W0Q - PubMed Abstract:
The mechanism of molecular oxygen entry into the buried active site of the copper amine oxidase family has been investigated in several family members using biochemical, structural and in silico methods. These studies have revealed a structurally conserved beta-sandwich which acts as a hydrophobic reservoir from which molecular oxygen can take several species-specific preferred pathways to the active site. Escherichia coli copper amine oxidase (ECAO) possesses an extra N-terminal domain that lies close to one entrance to the beta-sandwich. In order to investigate whether the presence of this domain alters molecular oxygen entry in this enzyme, xenon was used as a molecular oxygen binding-site probe. The resulting 2.5 A resolution X-ray crystal structure reveals xenon bound in similar positions to those observed in xenon-derivative crystal structures of other family members, suggesting that the N-terminal domain does not affect oxygen entry and that the E. coli enzyme takes up oxygen in a similar manner to the rest of the copper amine oxidase family.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds LS2 9JT, England.