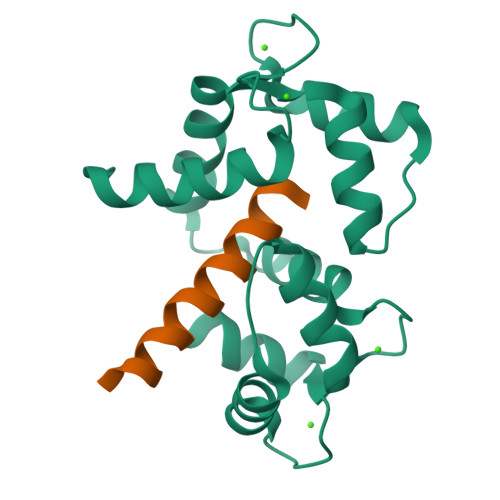
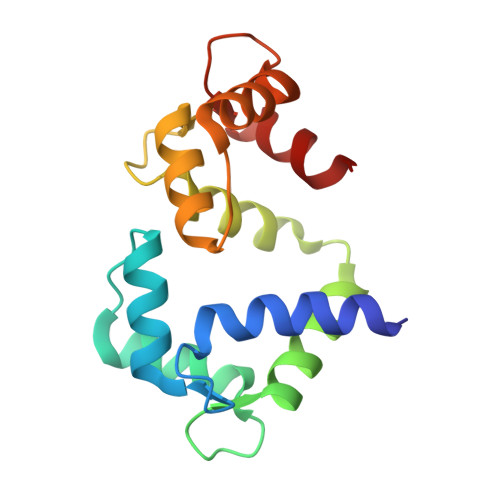
Determinants in Cav1 Channels that Regulate the Ca2+ Sensitivity of Bound Calmodulin.
Halling, D.B., Georgiou, D.K., Black, D.J., Yang, G., Fallon, J.L., Quiocho, F.A., Pedersen, S.E., Hamilton, S.L.(2009) J Biological Chem 284: 20041
- PubMed: 19473981
- DOI: https://doi.org/10.1074/jbc.M109.013326
- Primary Citation of Related Structures:
2VAY - PubMed Abstract:
Calmodulin binds to IQ motifs in the alpha(1) subunit of Ca(V)1.1 and Ca(V)1.2, but the affinities of calmodulin for the motif and for Ca(2+) are higher when bound to Ca(V)1.2 IQ. The Ca(V)1.1 IQ and Ca(V)1.2 IQ sequences differ by four amino acids. We determined the structure of calmodulin bound to Ca(V)1.1 IQ and compared it with that of calmodulin bound to Ca(V)1.2 IQ. Four methionines in Ca(2+)-calmodulin form a hydrophobic binding pocket for the peptide, but only one of the four nonconserved amino acids (His-1532 of Ca(V)1.1 and Tyr-1675 of Ca(V)1.2) contacts this calmodulin pocket. However, Tyr-1675 in Ca(V)1.2 contributes only modestly to the higher affinity of this peptide for calmodulin; the other three amino acids in Ca(V)1.2 contribute significantly to the difference in the Ca(2+) affinity of the bound calmodulin despite having no direct contact with calmodulin. Those residues appear to allow an interaction with calmodulin with one lobe Ca(2+)-bound and one lobe Ca(2+)-free. Our data also provide evidence for lobe-lobe interactions in calmodulin bound to Ca(V)1.2.
Organizational Affiliation:
Department of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, Texas 77030, USA.