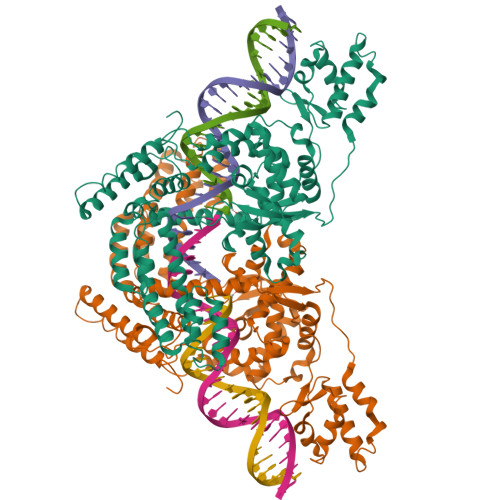
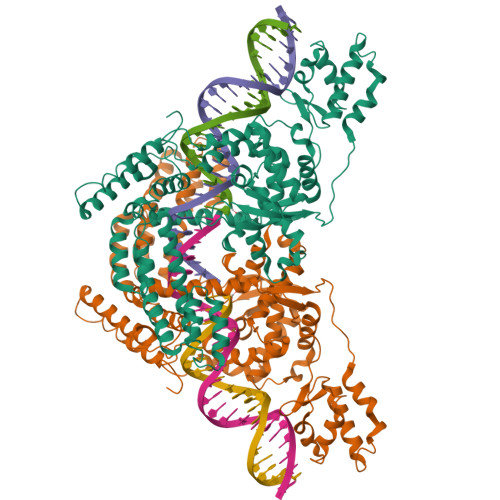
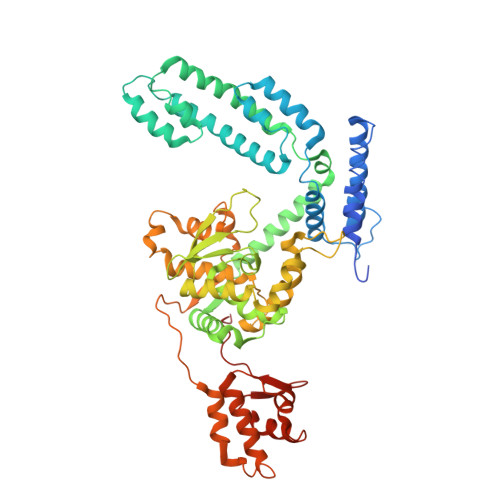
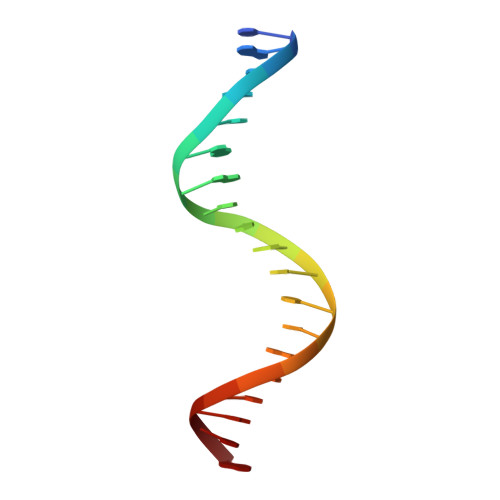
An Interlocked Dimer of the Protelomerase Telk Distorts DNA Structure for the Formation of Hairpin Telomeres
Aihara, H., Huang, W.M., Ellenberger, T.(2007) Mol Cell 27: 901
- PubMed: 17889664
- DOI: https://doi.org/10.1016/j.molcel.2007.07.026
- Primary Citation of Related Structures:
2V6E - PubMed Abstract:
The termini of linear chromosomes are protected by specialized DNA structures known as telomeres that also facilitate the complete replication of DNA ends. The simplest type of telomere is a covalently closed DNA hairpin structure found in linear chromosomes of prokaryotes and viruses. Bidirectional replication of a chromosome with hairpin telomeres produces a catenated circular dimer that is subsequently resolved into unit-length chromosomes by a dedicated DNA cleavage-rejoining enzyme known as a hairpin telomere resolvase (protelomerase). Here we report a crystal structure of the protelomerase TelK from Klebsiella oxytoca phage varphiKO2, in complex with the palindromic target DNA. The structure shows the TelK dimer destabilizes base pairing interactions to promote the refolding of cleaved DNA ends into two hairpin ends. We propose that the hairpinning reaction is made effectively irreversible by a unique protein-induced distortion of the DNA substrate that prevents religation of the cleaved DNA substrate.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110, USA.