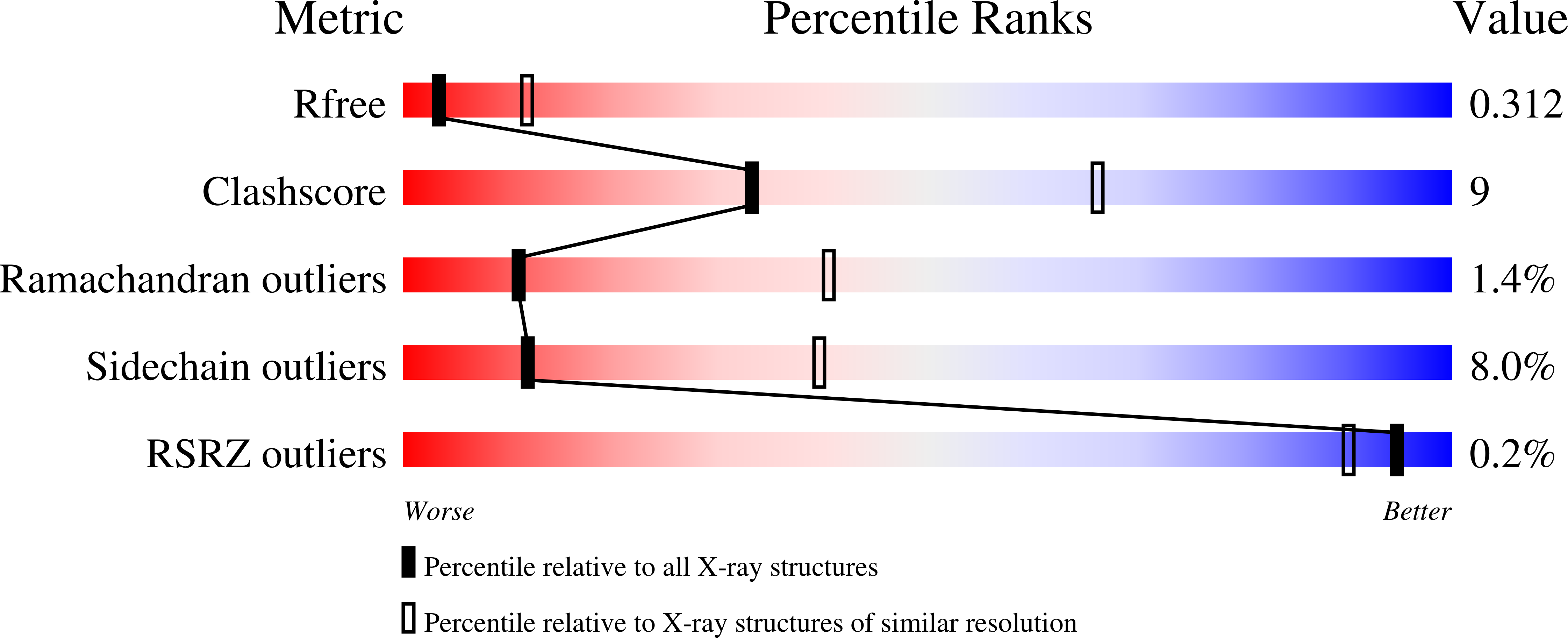
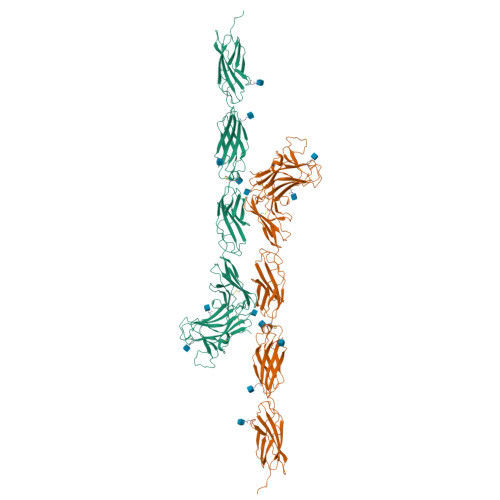
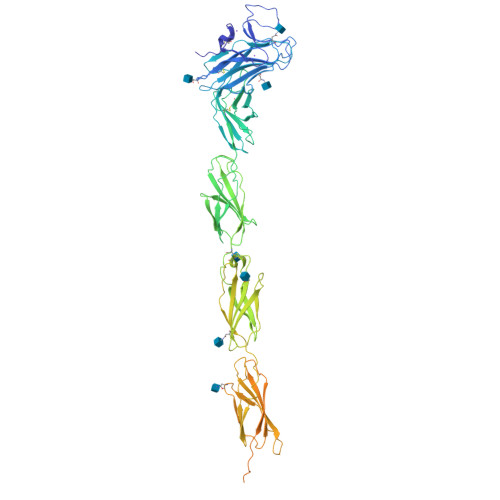Structure of a Tyrosine Phosphatase Adhesive Interaction Reveals a Spacer-Clamp Mechanism.
Aricescu, A.R., Siebold, C., Choudhuri, K., Chang, V.T., Lu, W., Davis, S.J., Van Der Merwe, P.A., Jones, E.Y.(2007) Science 317: 1217
- PubMed: 17761881
- DOI: https://doi.org/10.1126/science.1144646
- Primary Citation of Related Structures:
2V5Y - PubMed Abstract:
Cell-cell contacts are fundamental to multicellular organisms and are subject to exquisite levels of control. Human RPTPmu is a type IIB receptor protein tyrosine phosphatase that both forms an adhesive contact itself and is involved in regulating adhesion by dephosphorylating components of cadherin-catenin complexes. Here we describe a 3.1 angstrom crystal structure of the RPTPmu ectodomain that forms a homophilic trans (antiparallel) dimer with an extended and rigid architecture, matching the dimensions of adherens junctions. Cell surface expression of deletion constructs induces intercellular spacings that correlate with the ectodomain length. These data suggest that the RPTPmu ectodomain acts as a distance gauge and plays a key regulatory function, locking the phosphatase to its appropriate functional location.
Organizational Affiliation:
Cancer Research UK Receptor Structure Research Group, University of Oxford, Henry Wellcome Building of Genomic Medicine, Division of Structural Biology, Roosevelt Drive, Oxford OX3 7BN, UK.