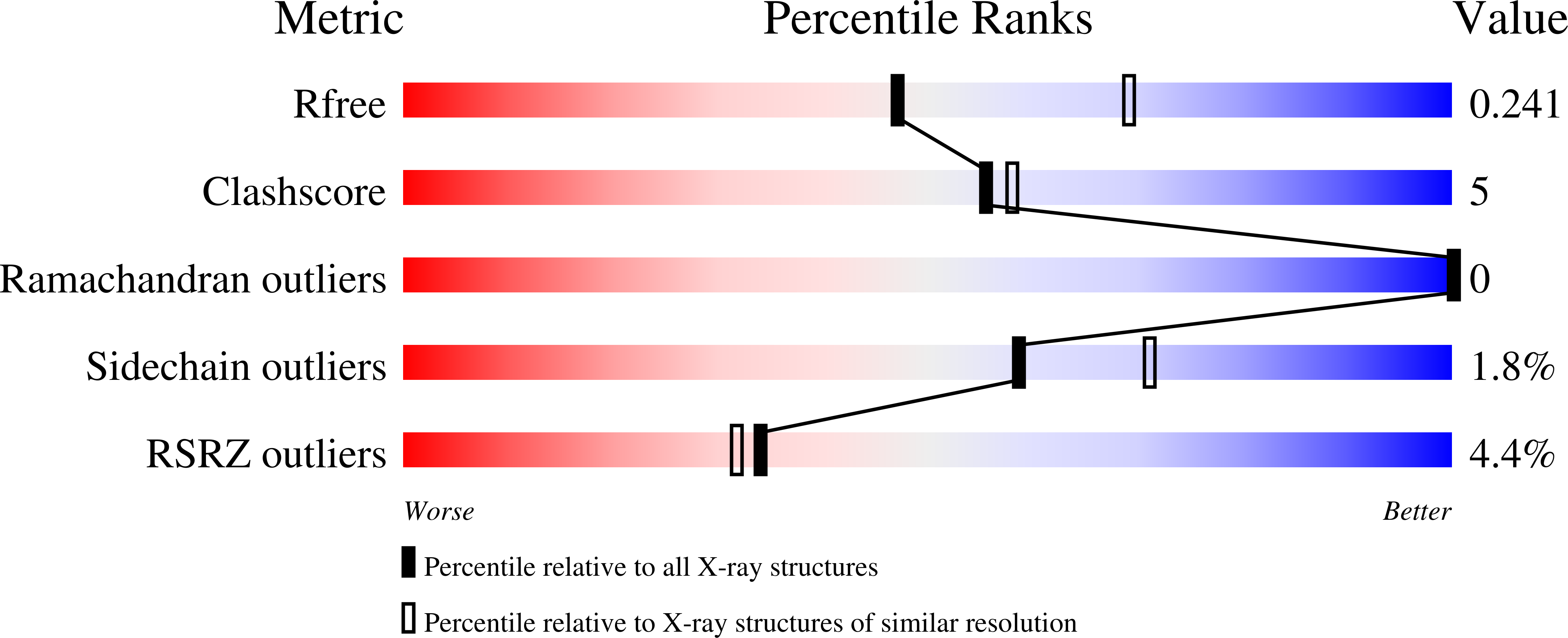
Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Hagelueken, G., Wiehlmann, L., Adams, T.M., Kolmar, H., Heinz, D.W., Tuemmler, B., Schubert, W.-D.(2007) Proc Natl Acad Sci U S A 104: 12276
- PubMed: 17636129
- DOI: https://doi.org/10.1073/pnas.0702919104
- Primary Citation of Related Structures:
2V3A, 2V3B - PubMed Abstract:
Crude oil spills represent a major ecological threat because of the chemical inertness of the constituent n-alkanes. The Gram-negative bacterium Pseudomonas aeruginosa is one of the few bacterial species able to metabolize such compounds. Three chromosomal genes, rubB, rubA1, and rubA2 coding for an NAD(P)H:rubredoxin reductase (RdxR) and two rubredoxins (Rdxs) are indispensable for this ability. They constitute an electron transport (ET) pathway that shuttles reducing equivalents from carbon metabolism to the membrane-bound alkane hydroxylases AlkB1 and AlkB2. The RdxR-Rdx system also is crucial as part of the oxidative stress response in archaea or anaerobic bacteria. The redox couple has been analyzed in detail as a model system for ET processes. We have solved the structure of RdxR of P. aeruginosa both alone and in complex with Rdx, without the need for cross-linking, and both structures were refined at 2.40- and 2.45-A resolution, respectively. RdxR consists of two cofactor-binding domains and a C-terminal domain essential for the specific recognition of Rdx. Only a small number of direct interactions govern mutual recognition of RdxR and Rdx, corroborating the transient nature of the complex. The shortest distance between the redox centers is observed to be 6.2 A.
Organizational Affiliation:
Molecular Host-Pathogen Interactions, Division of Structural Biology, Helmholtz Centre for Infection Research, Inhoffenstrasse 7, D-38124 Braunschweig, Germany.