Conformational Changes in the (Ala-84-Val) Mutant of Tumor Necrosis Factor
Saludjian, P., Prange, T., Kahn, R., Fourme, R., Tavernier, J., Van-Oostade, X., Fiers, W., Navaza, G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TUMOR NECROSIS FACTOR-ALPHA | 157 | Homo sapiens | Mutation(s): 1 | 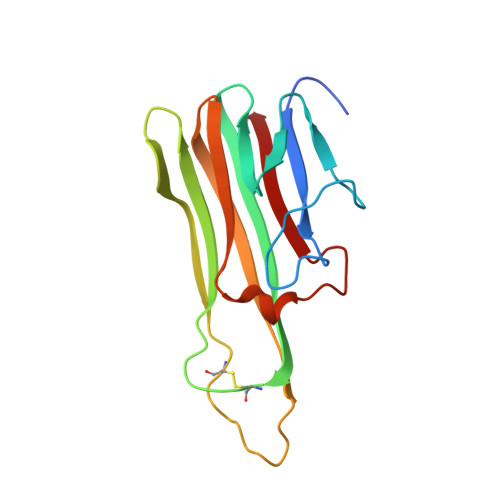 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01375 (Homo sapiens) Explore P01375 Go to UniProtKB: P01375 | |||||
PHAROS: P01375 GTEx: ENSG00000232810 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01375 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 166 | α = 90 |
| b = 166 | β = 90 |
| c = 93.7 | γ = 120 |
| Software Name | Purpose |
|---|---|
| SHARP | phasing |
| PROLSQ | refinement |