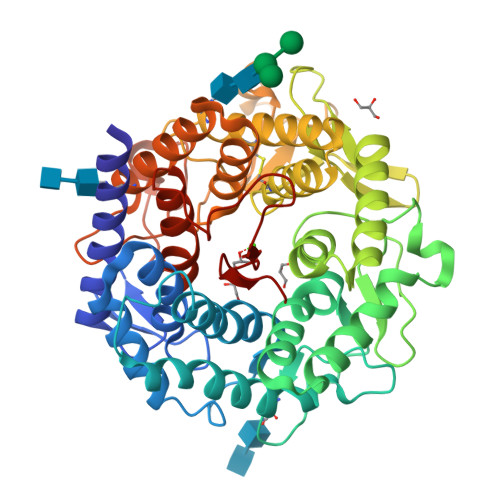
Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
Lobsanov, Y.D., Yoshida, T., Desmet, T., Nerinckx, W., Yip, P., Claeyssens, M., Herscovics, A., Howell, P.L.(2008) Acta Crystallogr D Biol Crystallogr 64: 227-236
- PubMed: 18323617
- DOI: https://doi.org/10.1107/S0907444907065572
- Primary Citation of Related Structures:
2RI8, 2RI9 - PubMed Abstract:
Class I alpha-mannosidases (glycoside hydrolase family GH47) play key roles in the maturation of N-glycans and the ER-associated degradation of unfolded glycoproteins. The 1.95 A resolution structure of a fungal alpha-1,2-mannosidase in complex with the substrate analogue methyl-alpha-D-lyxopyranosyl-(1',2)-alpha-D-mannopyranoside (LM) shows the intact disaccharide spanning the -1/+1 subsites, with the D-lyxoside ring in the -1 subsite in the 1C4 chair conformation, and provides insight into the mechanism of catalysis. The absence of the C5' hydroxymethyl group on the D-lyxoside moiety results in the side chain of Arg407 adopting two alternative conformations: the minor one interacting with Asp375 and the major one interacting with both the D-lyxoside and the catalytic base Glu409, thus disrupting its function. Chemical modification of Asp375 has previously been shown to inactivate the enzyme. Taken together, the data suggest that Arg407, which belongs to the conserved sequence motif RPExxE, may act to modulate the activity of the enzyme. The proposed mechanism for modulating the activity is potentially a general mechanism for this superfamily.
Organizational Affiliation:
Program in Molecular Structure and Function, Research Institute, The Hospital for Sick Children, 555 University Avenue, Toronto, Ontario M5G 1X8, Canada.