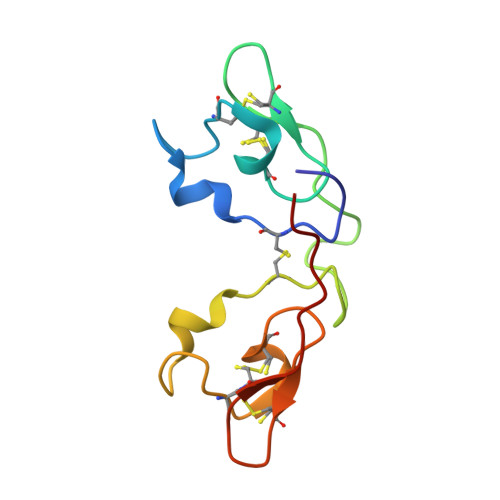
Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Moulaei, T., Botos, I., Ziolkowska, N.E., Bokesch, H.R., Krumpe, L.R., McKee, T.C., O'Keefe, B.R., Dauter, Z., Wlodawer, A.(2007) Protein Sci 16: 2756-2760
- PubMed: 17965185
- DOI: https://doi.org/10.1110/ps.073157507
- Primary Citation of Related Structures:
2QSK, 2QT4 - PubMed Abstract:
The crystal structures of the natural and recombinant antiviral lectin scytovirin (SVN) were solved by single-wavelength anomalous scattering and refined with data extending to 1.3 A and 1.0 A resolution, respectively. A molecule of SVN consists of a single chain 95 amino acids long, with an almost perfect sequence repeat that creates two very similar domains (RMS deviation 0.25 A for 40 pairs of Calpha atoms). The crystal structure differs significantly from a previously published NMR structure of the same protein, with the RMS deviations calculated separately for the N- and C-terminal domains of 5.3 A and 3.7 A, respectively, and a very different relationship between the two domains. In addition, the disulfide bonding pattern of the crystal structures differs from that described in the previously published mass spectrometry and NMR studies.
Organizational Affiliation:
Protein Structure Section, Macromolecular Crystallography Laboratory, National Cancer Institute, NCI-Frederick, Frederick, Maryland 21702-1201, USA.