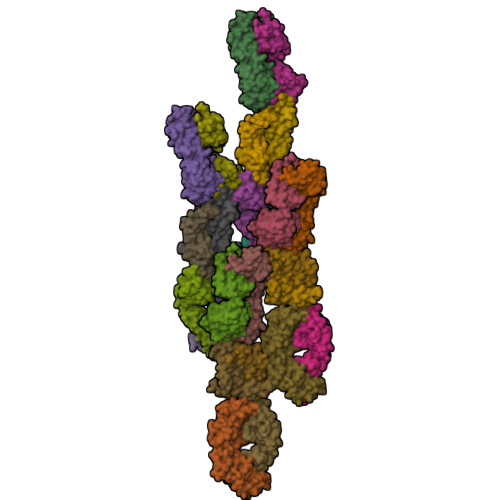
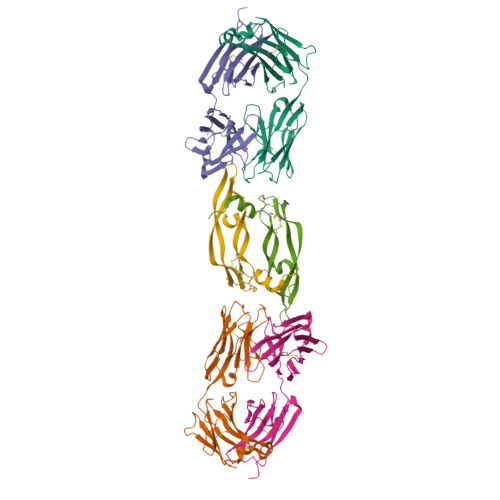
High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries
Fellouse, F.A., Esaki, K., Birtalan, S., Raptis, D., Cancasci, V.J., Koide, A., Jhurani, P., Vasser, M., Wiesmann, C., Kossiakoff, A.A., Koide, S., Sidhu, S.S.(2007) J Mol Biology 373: 924-940
- PubMed: 17825836
- DOI: https://doi.org/10.1016/j.jmb.2007.08.005
- Primary Citation of Related Structures:
2QR0 - PubMed Abstract:
We have previously established a minimalist approach to antibody engineering by using a phage-displayed framework to support complementarity determining region (CDR) diversity restricted to a binary code of tyrosine and serine. Here, we systematically augmented the original binary library with additional levels of diversity and examined the effects. The diversity of the simplest library, in which only heavy chain CDR positions were randomized by the binary code, was expanded in a stepwise manner by adding diversity to the light chain, by diversifying non-paratope residues that may influence CDR conformations, and by adding additional chemical diversity to CDR-H3. The additional diversity incrementally improved the affinities of antibodies raised against human vascular endoethelial growth factor and the structure of an antibody-antigen complex showed that tyrosine side-chains are sufficient to mediate most of the interactions with antigen, but a glycine residue in CDR-H3 was critical for providing a conformation suitable for high-affinity binding. Using new high-throughput procedures and the most complex library, we produced multiple high-affinity antibodies with dissociation constants in the single-digit nanomolar range against a wide variety of protein antigens. Thus, this fully synthetic, minimalist library has essentially recapitulated the capacity of the natural immune system to generate high-affinity antibodies. Libraries of this type should be highly useful for proteomic applications, as they minimize inherent complexities of natural antibodies that have hindered the establishment of high-throughput procedures. Furthermore, analysis of a large number of antibodies derived from these well-defined and simplistic libraries allowed us to uncover statistically significant trends in CDR sequences, which provide valuable insights into antibody library design and into factors governing protein-protein interactions.
Organizational Affiliation:
Department of Protein Engineering, Genentech Inc, 1 DNA Way, South San Francisco, CA 94080, USA.