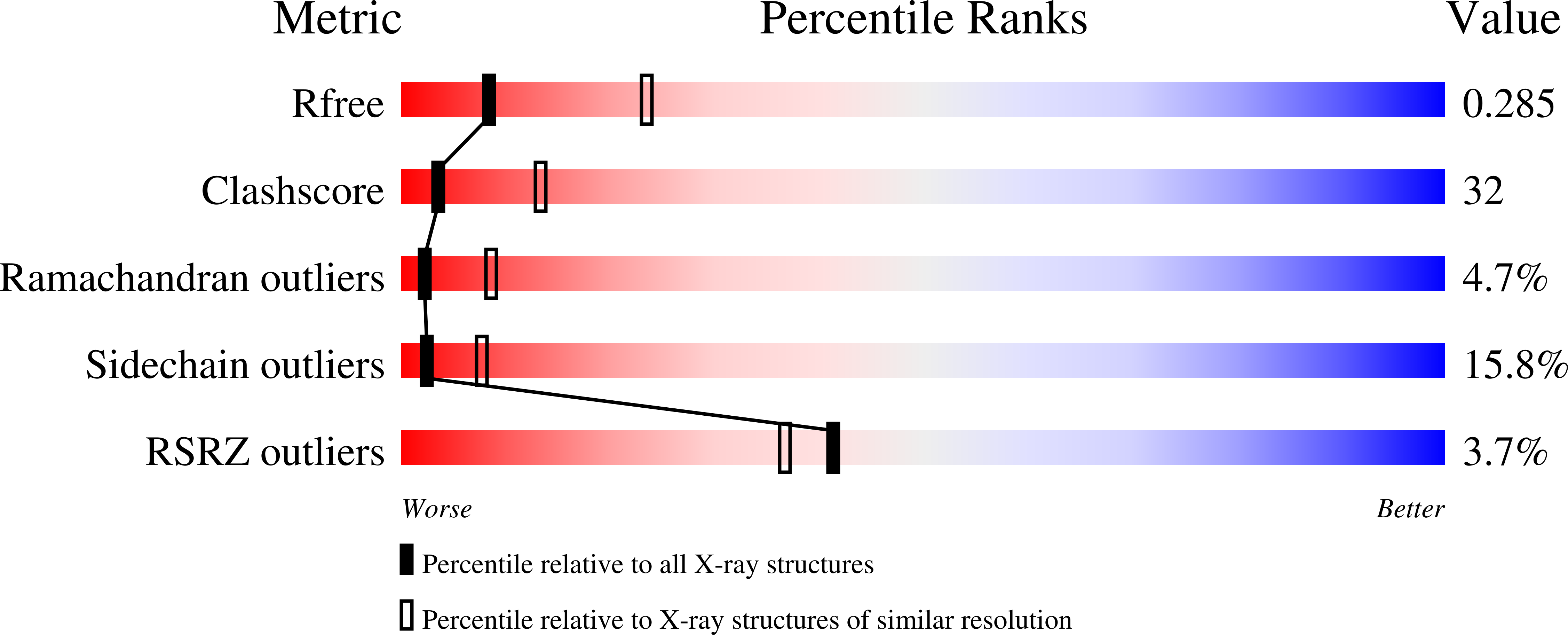
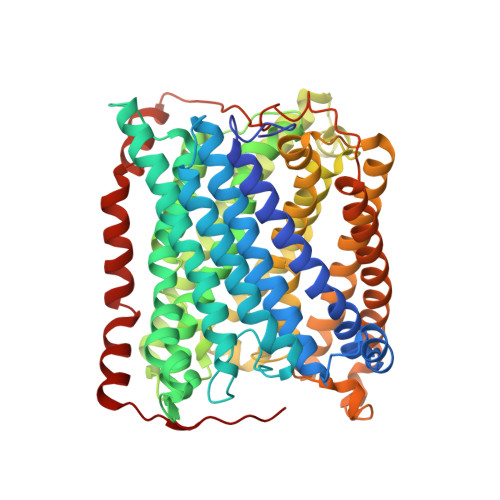
An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
Liu, B., Luna, V.M., Chen, Y., Stout, C.D., Fee, J.A.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 1029-1034
- PubMed: 18084085
- DOI: https://doi.org/10.1107/S1744309107054176
- Primary Citation of Related Structures:
2QPD, 2QPE - PubMed Abstract:
Past work has shown that it is feasible to mutate surface residues of soluble proteins and to a lesser extent membrane proteins in order to improve their crystallization behavior. Described here is a successful application of this approach to the integral membrane protein Thermus thermophilus cytochrome ba(3) oxidase. Two mutant forms of this enzyme (I-K258R and I-K258R/II-E4Q) were created in which symmetrical crystal contacts within crystals of wild-type enzyme were modified. These mutant proteins had greatly shortened crystallization times, decreasing from approximately 30 d for the wild type to 1-3 d for the mutants, and crystallization was highly reproducible. Native-like proteins crystallize in space group P4(3)2(1)2, whereas the mutant proteins crystallize in space group P4(1)2(1)2 with a different packing arrangement. Crystals of the P4(3)2(1)2 form occasionally diffracted to 2.4-2.3 A resolution following controlled dehydration, while those of the P4(1)2(1)2 form routinely diffracted to between 3.0 and 2.6 A for crystals that had been cryoprotected but not dehydrated.
Organizational Affiliation:
The Scripps Research Institute, Department of Molecular Biology, MB-8, 10550 North Torrey Pines Road, La Jolla CA 92037, USA.