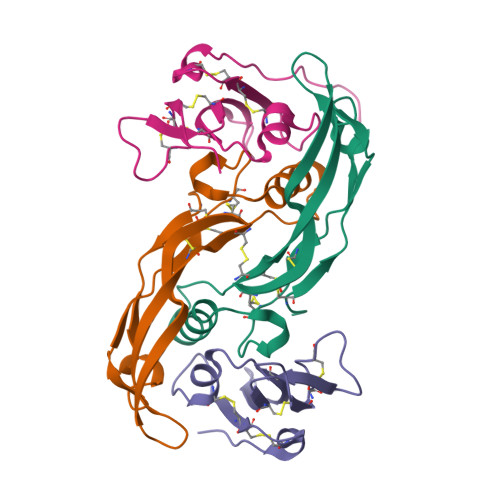
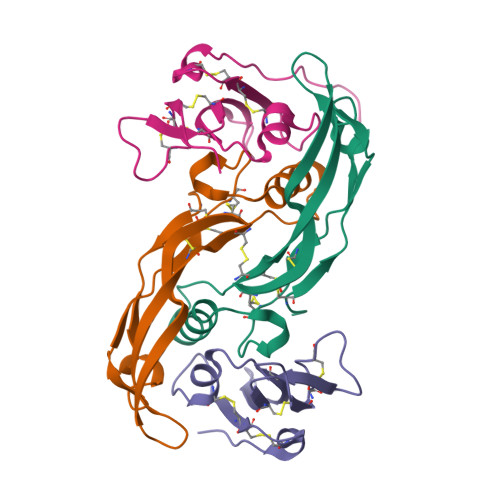
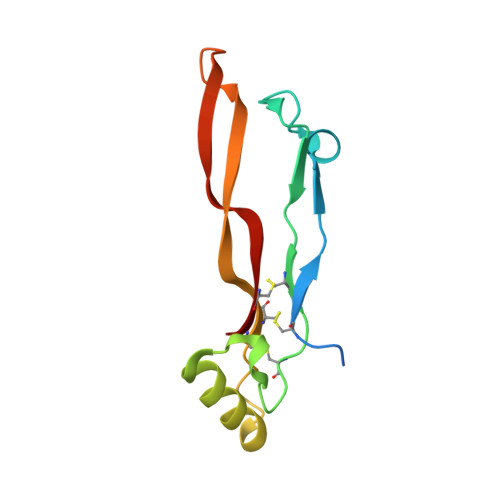
Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome.
Kotzsch, A., Nickel, J., Seher, A., Heinecke, K., van Geersdaele, L., Herrmann, T., Sebald, W., Mueller, T.D.(2008) J Biological Chem 283: 5876-5887
- PubMed: 18160401
- DOI: https://doi.org/10.1074/jbc.M706029200
- Primary Citation of Related Structures:
2QJ9, 2QJA, 2QJB - PubMed Abstract:
Bone morphogenetic proteins regulate many developmental processes during embryogenesis as well as tissue homeostasis in the adult. Signaling of bone morphogenetic proteins (BMPs) is accomplished by binding to two types of serine/threonine kinase transmembrane receptors termed type I and type II. Because a large number of ligands signal through a limited number of receptors, ligand-receptor interaction in the BMP superfamily is highly promiscuous, with a ligand binding to various receptors and a receptor binding many different BMP ligands. In this study we investigate the interaction of BMP-2 with its two high affinity type I receptors, BMP receptors IA (BMPR-IA) and BMPR-IB. Interestingly, 50% of the residues in the BMP-2 binding epitope of the BMPR-IA receptor are exchanged in BMPR-IB without a decrease in binding affinity or specificity for BMP-2. Our structural and functional analyses show that promiscuous binding of BMP-2 to both type I receptors is achieved by inherent backbone and side-chain flexibility as well as by variable hydration of the ligand-receptor interface enabling the BMP-2 surface to adapt to different receptor geometries. Despite the high degree of amino acid variability found in BMPR-IA and BMPR-IB binding equally to BMP-2, three single point missense mutations in the ectodomain of BMPR-IA cannot be tolerated. In juvenile polyposis syndrome these mutations have been shown to inactivate BMPR-IA. On the basis of our biochemical and biophysical analyses, we can show that the mutations, which are located outside the ligand binding epitope, alter the local or global fold of the receptor, thereby inactivating BMPR-IA and causing a loss of the BMP-2 tumor suppressor function in colon epithelial cells.
Organizational Affiliation:
Lehrstuhl für Botanik I-Molekulare Pflanzenphysiologie und Biophysik, Julius-von-Sachs-Institut der Universität Würzburg, Julius-von-Sachs-Platz 2, D-97082 Würzburg, Germany.