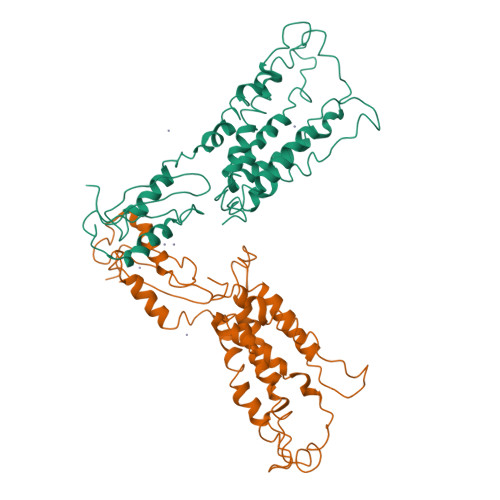
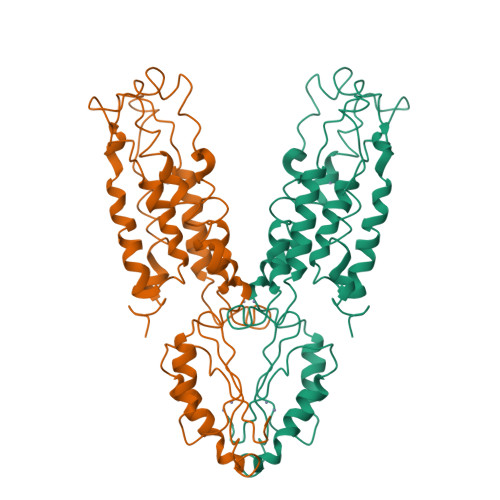
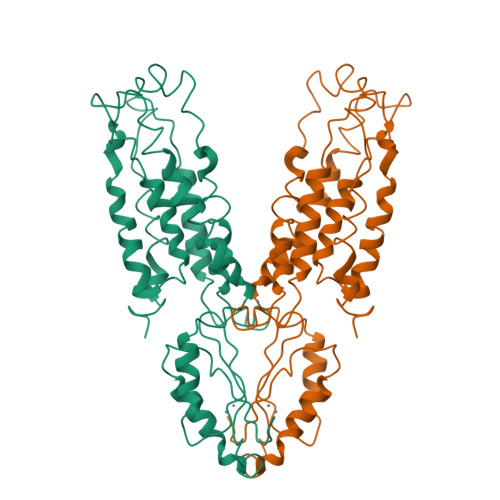
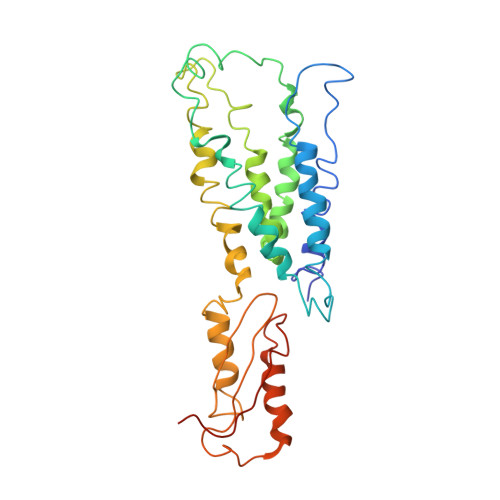
Structure of the zinc transporter YiiP.
Lu, M., Fu, D.(2007) Science 317: 1746-1748
- PubMed: 17717154
- DOI: https://doi.org/10.1126/science.1143748
- Primary Citation of Related Structures:
2QFI - PubMed Abstract:
YiiP is a membrane transporter that catalyzes Zn2+/H+ exchange across the inner membrane of Escherichia coli. Mammalian homologs of YiiP play critical roles in zinc homeostasis and cell signaling. Here, we report the x-ray structure of YiiP in complex with zinc at 3.8 angstrom resolution. YiiP is a homodimer held together in a parallel orientation through four Zn2+ ions at the interface of the cytoplasmic domains, whereas the two transmembrane domains swing out to yield a Y-shaped structure. In each protomer, the cytoplasmic domain adopts a metallochaperone-like protein fold; the transmembrane domain features a bundle of six transmembrane helices and a tetrahedral Zn2+ binding site located in a cavity that is open to both the membrane outer leaflet and the periplasm.
Organizational Affiliation:
Department of Biology, Brookhaven National Laboratory, Upton, NY 11973, USA.