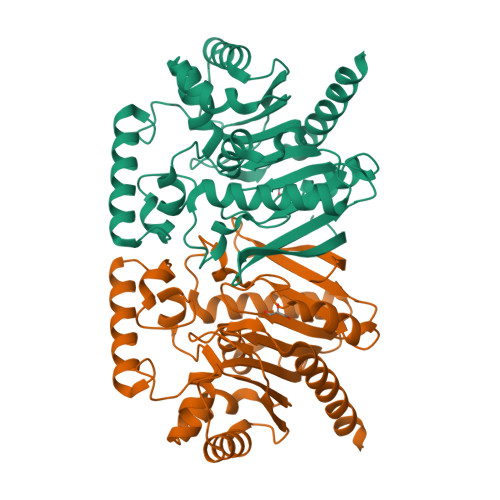
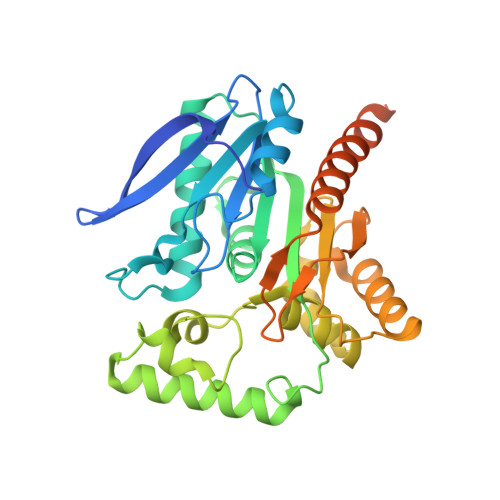
Structure of a Trypanosoma brucei alpha/beta-hydrolase fold protein with unknown function.
Merritt, E.A., Holmes, M., Buckner, F.S., Van Voorhis, W.C., Quartly, E., Phizicky, E.M., Lauricella, A., Luft, J., DeTitta, G., Neely, H., Zucker, F., Hol, W.G.(2008) Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 474-478
- PubMed: 18540054
- DOI: https://doi.org/10.1107/S174430910801141X
- Primary Citation of Related Structures:
2Q0X - PubMed Abstract:
The structure of a structural genomics target protein, Tbru020260AAA from Trypanosoma brucei, has been determined to a resolution of 2.2 A using multiple-wavelength anomalous diffraction at the Se K edge. This protein belongs to Pfam sequence family PF08538 and is only distantly related to previously studied members of the alpha/beta-hydrolase fold family. Structural superposition onto representative alpha/beta-hydrolase fold proteins of known function indicates that a possible catalytic nucleophile, Ser116 in the T. brucei protein, lies at the expected location. However, the present structure and by extension the other trypanosomatid members of this sequence family have neither sequence nor structural similarity at the location of other active-site residues typical for proteins with this fold. Together with the presence of an additional domain between strands beta6 and beta7 that is conserved in trypanosomatid genomes, this suggests that the function of these homologs has diverged from other members of the fold family.
Organizational Affiliation:
Structural Genomics of Pathogenic Protozoa (SGPP) Consortium, USA. merritt@u.washington.edu