Structures of 5-methylthioribose kinase reveal substrate specificity and unusual mode of nucleotide binding
Ku, S.-Y., Yip, P., Cornell, K.A., Riscoe, M.K., Behr, J.-B., Guillerm, G., Howell, P.L.(2007) J Biological Chem 282: 22195-22206
- PubMed: 17522047
- DOI: https://doi.org/10.1074/jbc.M611045200
- Primary Citation of Related Structures:
2PU8, 2PUI, 2PUL, 2PUN, 2PUP - PubMed Abstract:
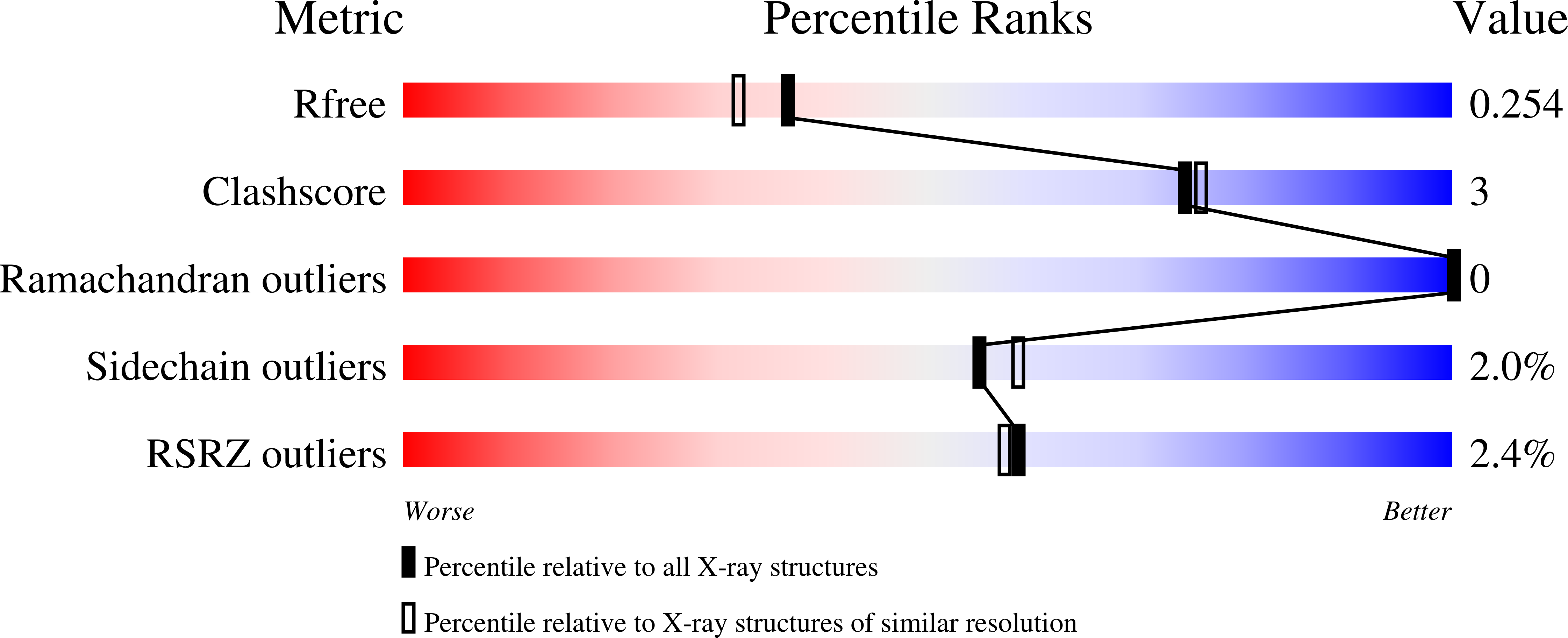
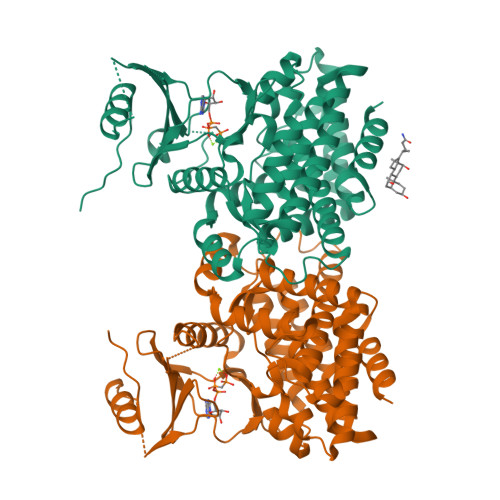
The methionine salvage pathway is ubiquitous in all organisms, but metabolic variations exist between bacteria and mammals. 5-Methylthioribose (MTR) kinase is a key enzyme in methionine salvage in bacteria and the absence of a mammalian homolog suggests that it is a good target for the design of novel antibiotics. The structures of the apo-form of Bacillus subtilis MTR kinase, as well as its ADP, ADP-PO(4), AMPPCP, and AMPPCP-MTR complexes have been determined. MTR kinase has a bilobal eukaryotic protein kinase fold but exhibits a number of unique features. The protein lacks the DFG motif typically found at the beginning of the activation loop and instead coordinates magnesium via a DXE motif (Asp(250)-Glu(252)). In addition, the glycine-rich loop of the protein, analogous to the "Gly triad" in protein kinases, does not interact extensively with the nucleotide. The MTR substrate-binding site consists of Asp(233) of the catalytic HGD motif, a novel twin arginine motif (Arg(340)/Arg(341)), and a semi-conserved W-loop, which appears to regulate MTR binding specificity. No lobe closure is observed for MTR kinase upon substrate binding. This is probably because the enzyme lacks the lobe closure/inducing interactions between the C-lobe of the protein and the ribosyl moiety of the nucleotide that are typically responsible for lobe closure in protein kinases. The current structures suggest that MTR kinase has a dissociative mechanism.
Organizational Affiliation:
Program in Molecular Structure and Function, Research Institute, Hospital for Sick Children, Toronto, Ontario, Canada.