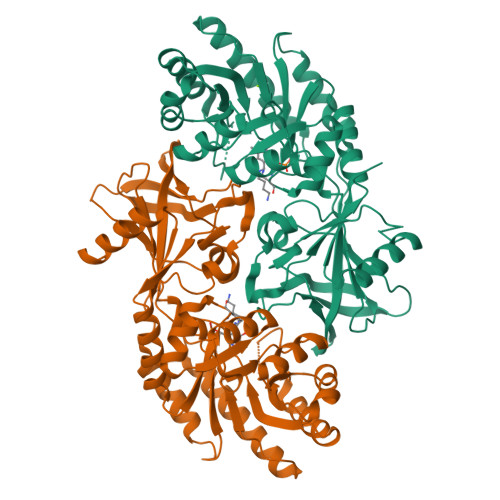
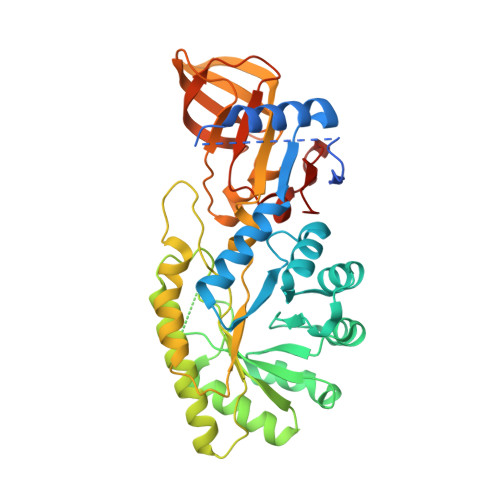
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases.
Lee, J., Michael, A.J., Martynowski, D., Goldsmith, E.J., Phillips, M.A.(2007) J Biological Chem 282: 27115-27125
- PubMed: 17626020
- DOI: https://doi.org/10.1074/jbc.M704066200
- Primary Citation of Related Structures:
2PLJ, 2PLK - PubMed Abstract:
The beta/alpha-barrel fold type basic amino acid decarboxylases include eukaryotic ornithine decarboxylases (ODC) and bacterial and plant enzymes with activity on L-arginine and meso-diaminopimelate. These enzymes catalyze essential steps in polyamine and lysine biosynthesis. Phylogenetic analysis suggests that diverse bacterial species also contain ODC-like enzymes from this fold type. However, in comparison with the eukaryotic ODCs, amino acid differences were identified in the sequence of the 3(10)-helix that forms a key specificity element in the active site, suggesting they might function on novel substrates. Putative decarboxylases from a phylogenetically diverse range of bacteria were characterized to determine their substrate preference. Enzymes from species within Methanosarcina, Pseudomonas, Bartonella, Nitrosomonas, Thermotoga, and Aquifex showed a strong preference for L-ornithine, whereas the enzyme from Vibrio vulnificus (VvL/ODC) had dual specificity functioning well on both L-ornithine and L-lysine. The x-ray structure of VvL/ODC was solved in the presence of the reaction products putrescine and cadaverine to 1.7 and 2.15A, respectively. The overall structure is similar to eukaryotic ODC; however, reorientation of the 3(10)-helix enlarging the substrate binding pocket allows L-lysine to be accommodated. The structure of the putrescine-bound enzyme suggests that a bridging water molecule between the shorter L-ornithine and key active site residues provides the structural basis for VvL/ODC to also function on this substrate. Our data demonstrate that there is greater structural and functional diversity in bacterial polyamine biosynthetic decarboxylases than previously suspected.
Organizational Affiliation:
Departments of Pharmacology, University of Texas Southwestern Medical Center, Dallas, Texas 75390-9041.