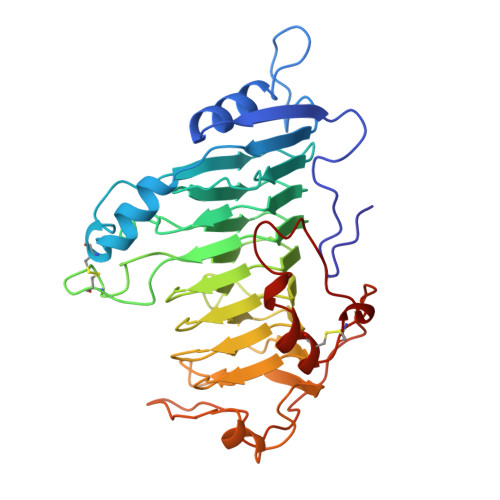
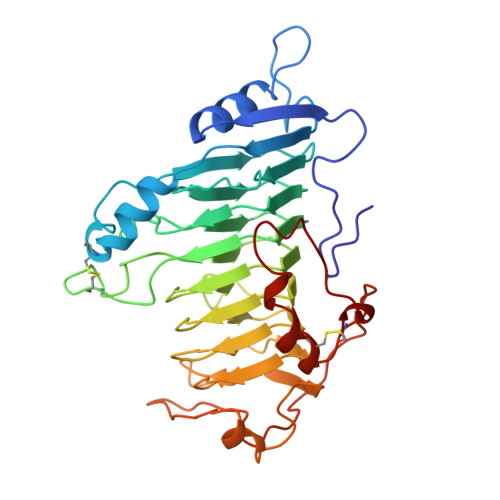
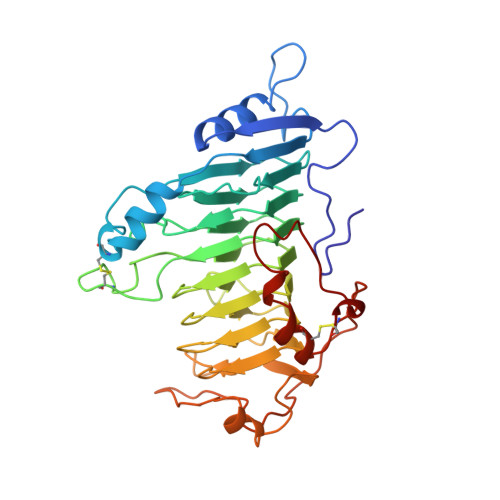
Protein motifs. 3. The parallel beta helix and other coiled folds.
Yoder, M.D., Jurnak, F.(1995) FASEB J 9: 335-342
- PubMed: 7896002
- DOI: https://doi.org/10.1096/fasebj.9.5.7896002
- Primary Citation of Related Structures:
2PEC - PubMed Abstract:
A new type of structural domain, composed of all parallel beta strands, has been observed within the last year. An analysis of the basic types suggests that there are two distinct classes: the parallel beta helices, which belong to a tri beta-strand category, and the beta roll, which belongs to a di beta-strand category. The novel structural features of each class are described and the proteins belonging to each category are summarized. Proteins with the parallel beta helix fold include three pectate lyases and the tailspike protein from P22 phage. Proteins with the beta roll fold include two alkaline proteases. Although the parallel beta composition is emphasized, the same set of proteins share another common structural feature with several other proteins containing alpha helices: the polypeptide backbone is folded into a coiled structure in which each coil has the same 3-dimensional arrangement of a group of secondary structural elements. In addition to parallel beta domains, the other groups include the alpha/beta coiled fold, as represented by ribonuclease inhibitor, and the alpha/alpha coiled fold, as represented by lipovitellin and soluble lytic transglycoslyase. Novel features of the alpha/beta and alpha/alpha coiled folds are summarized.
Organizational Affiliation:
School of Biological Sciences, Division of Cell Biology and Biophysics, University of Missouri, Kansas City 64110.