PGH2 degradation pathway catalyzed by GSH-heme complex bound microsomal prostaglandin E2 synthase type 2: the first example of a dual-function enzyme.
Yamada, T., Takusagawa, F.(2007) Biochemistry 46: 8414-8424
- PubMed: 17585783
- DOI: https://doi.org/10.1021/bi700605m
- Primary Citation of Related Structures:
2PBJ - PubMed Abstract:
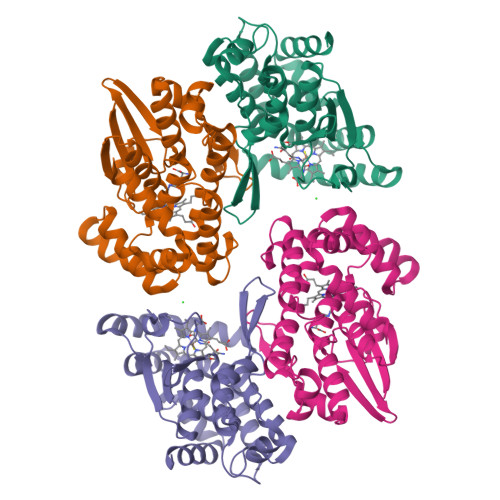
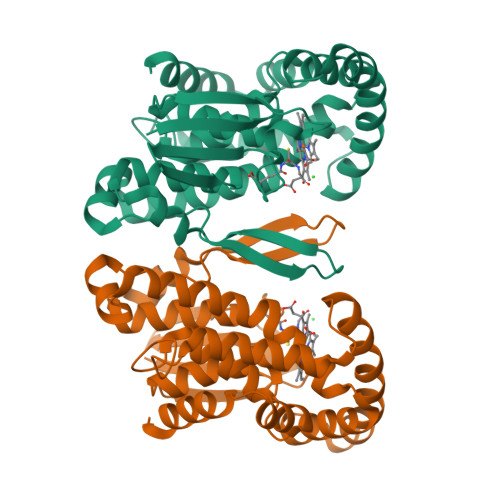
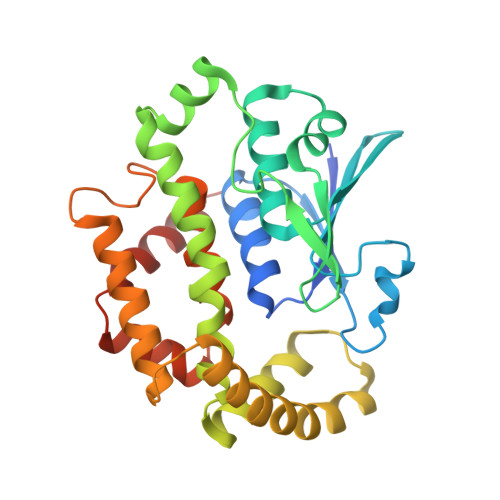
Prostaglandin E2 synthase (PGES) catalyzes the isomerization of PGH2 to PGE2. PGES type 2 (mPGES-2) is a membrane-associated enzyme, whose N-terminal section is apparently inserted into the lipid bilayer. Both intact and N-terminal truncated enzymes have been isolated and have similar catalytic activity. The recombinant N-terminal truncated enzyme purified from Escherichia coli HB101 grown in LB medium containing delta-aminolevulinate and Fe(NO3)3 has a red color, while the same enzyme purified from the same E. coli grown in minimal medium has no color. The red-colored enzyme has been characterized by mass, fluorescence, and EPR spectroscopies and X-ray crystallography. The enzyme is found to contain bound glutathione (GSH) and heme. GSH binds to the active site with six H-bonds, while a heme is complexed with bound GSH forming a S-Fe coordination bond with no polar interaction with mPGES-2. There is a large open space between the heme and the protein, where a PGH2 might be able to bind. The heme dissociation constant is 0.53 microM, indicating that mPGES-2 has relatively strong heme affinity. Indeed, expression of mPGES-2 in E. coli stimulates heme biosynthesis. Although mPGES-2 has been reported to be a GSH-independent PGES, the crystal structure and sequence analysis indicate that mPGES-2 is a GSH-binding protein. The GSH-heme complex-bound enzyme (mPGES-2h) catalyzes formation of 12(S)-hydroxy-5(Z),8(E),10(E)-heptadecatrienoic acid and malondialdehyde from PGH2, but not formation of PGE2. The following kinetic parameters at 37 degrees C were determined: KM = 56 microM, kcat = 63 s-1, and kcat/KM = 1.1 x 10(6) M-1 s-1. They suggest that mPGES-2h has significant catalytic activity for PGH2 degradation. It is possible that both GSH-heme complex-free and -bound enzymes are present in the same tissues. mPGES-2 in heme-rich liver is most likely to become the form of mPGES-2h and might be involved in degradation reactions similar to that of cytochrome P450. Since mPGES-2 is an isomerase and mPGES-2h is a lyase, mPGES-2 cannot simply be classified into one of six classes set by the International Union of Biochemistry and Molecular Biology.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, 1200 Sunnyside Avenue, Lawrence, Kansas 66045-7534, USA.